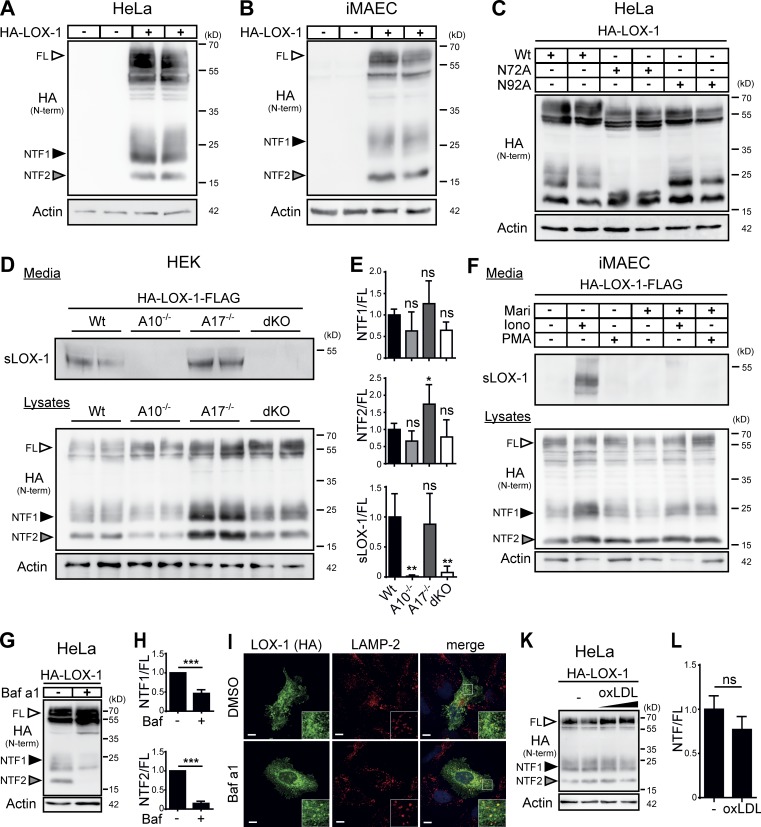
Figure 1.
LOX-1 NTFs are generated by ADAM10 and lysosomal proteases. (A and B) Two LOX-1 NTFs were detected by Western blotting upon overexpression of HA-LOX-1 in either HeLa (A) or iMAEC (B) cells. (C) Usage of the potential glycosylation sites at N72 or N92 was analyzed in HeLa cells using respective mutants. (D) HEK cells deficient for ADAM10, ADAM17, or both and WT cells were transfected with HA-LOX-1-FLAG. sLOX-1 was recovered from conditioned media after 16 h by TCA precipitation. (E) Quantification of D. N = 2–3, n = 4–6. One-way ANOVA with Dunnett’s post hoc test. (F) iMAECs stably expressing HA-LOX-1-FLAG were preincubated for 2 h in serum-free DMEM containing 10 µM marimastat (Mari) or DMSO. Cells were treated for 30 min with 1 µM ionomycin (Iono) or 100 nM PMA. Cell lysates and conditioned media were analyzed as in D. (G) Lysosomal processing of HA-LOX-1 was blocked in HeLa cells by incubation with 100 nM bafilomycin a1 (Baf a1) for 24 h. (H) Quantification of G. N = 3, n = 3. Student’s t test. (I) Delivery of HA-LOX-1 to LAMP-2–positive compartments was visualized by indirect immunofluorescence in HeLa cells treated with either DMSO or 100 nM bafilomycin a1 for 6 h. Bars, 10 µm. (K) HA-LOX-1–expressing HeLa cells were treated for 4 h with 40 or 80 µg/ml oxLDL, and NTF formation was analyzed by Western blotting. (L) Quantification of NTF1+2/FL ratios of cells treated with 40 µg/ml oxLDL. N = 2, n = 4. Student’s t test. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant. N, the number of independent experiments; n, the number of individual samples for quantification. All data are shown as mean ± SD.