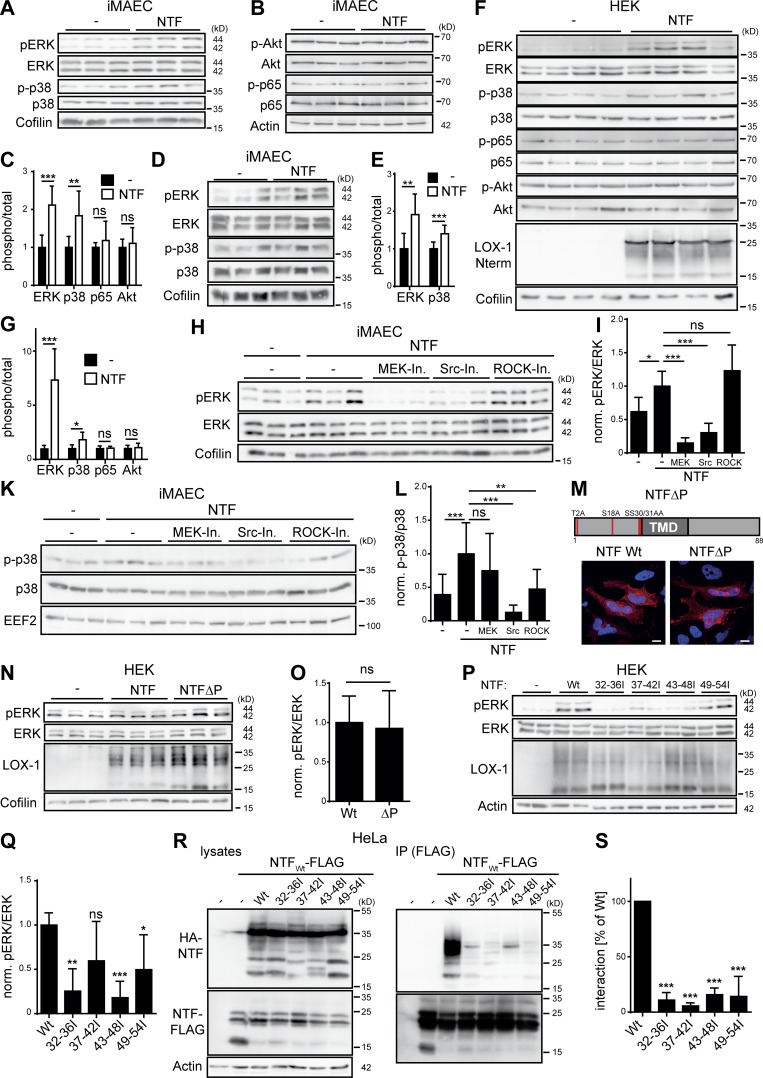
Figure 6.
The LOX-1 NTF autonomously activates MAP kinases. (A and B) iMAECs were stably transduced with the pMSCV puro vector (–) or a LOX-1-NTF coding construct. Levels of phosphorylated as well as total ERK1/2, p38 (A), p65, and Akt (B) were analyzed by Western blotting. (C) Quantification of A and B. N = 3, n = 9. Student’s t test. (D) Control or NTF-transduced iMAECs were starved for 16 h in serum-free DMEM and subsequently analyzed for activation of MAP kinases. (E) Quantification of D. N = 2, n = 6. Student’s t test. (F) Vector (–) or LOX-1 NTF transfected HEK cells were selected with puromycin for 4 d. (G) Quantification of F. N = 2, n = 7. Student’s t test. (H) Empty vector (–) or LOX-1 NTF transduced iMAECs were treated for 3 h with 25 µM U0126 (MEK-Inh.), 1 µM Saracatinib (Src-Inh.), or 10 µM Y-27632 (ROCK-Inh.) and subsequently analyzed for ERK1/2 activation. (I) Quantification of H. N = 2, n = 6. One-way ANOVA with Tukey’s post hoc test. (K) Pathways upstream of p38 MAP kinase activation were assessed as depicted in H. (L) Quantification of K. n = 4, n = 12. One-way ANOVA with Tukey’s post hoc test. (M) Subcellular sorting of an unphosphorylatable LOX-1 NTF (NTFΔP) was compared with the WT NTF by indirect immunofluorescence in HeLa cells. Bars, 10 µm. (N) HEK cells were transfected and incubated for 4 d with 10 µg/ml puromycin. Activation of ERK1/2 was monitored by Western blotting. (O) Quantification of N. pERK/ERK levels were normalized (norm.) to LOX-1 NTF expression. N = 2, n = 6. Student’s t test. (P) Induction of pERK by LOX-1 TMD mutants was analyzed by Western blotting. (Q) pERK/ERK ratios were normalized to NTF expression. N = 3, n = 6. One-way ANOVA with Dunnett’s post hoc test. (R) Coimmunoprecipitation of the HA-tagged NTF mutants with a FLAG-tagged WT LOX-1 NTF from lysates of transfected HEK cells. (S) Quantification from N = 3, n = 3 experiments. One-way ANOVA with Dunnett’s post hoc test. As described in Materials and methods, phosphorylated and total forms of ERK, p38, p65, and Akt were detected from the same membranes. After detection of the phosphorylated forms, membranes were stripped and reprobed to detect the total proteins. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant. N, the number of independent experiments; n, the number of individual samples for quantification. All data are shown as mean ± SD.