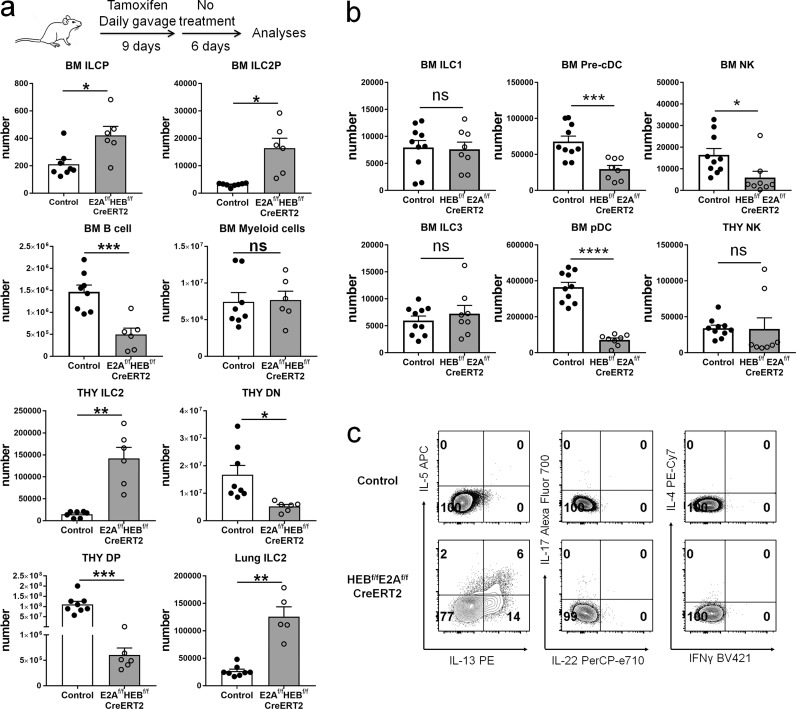
Figure 6.
Inducible ablation of E proteins promotes ILC2 differentiation in vivo. (a) ROSA26-CreERT2; E2Af/f; HEBf/f and control (E2Af/f; HEBf/f or ROSA26-CreERT2) mice were treated to induce the deletion of E proteins as diagrammed on the top. The definitions of the BM populations are ILCP (Lin−Thy1.2+IL-7+ST2−Sca-1−cKit+α4β7+PD-1+), ILC2P (Lin−Thy1.2+IL-7+ST2+Sca-1+CD25+α4β7+), B cells (B220+CD19+), and myeloid cells (Mac-1+). ILC2s in the thymus and lung are Lin−Thy1.2+ST2+. Total cell numbers of each indicated subset were quantified. Data shown are pooled from three experiments (n = 5–8). (b) Lineage cocktails for BM ILC1, BM ILC3, and BM NK include antibodies against FcεRI, B220, CD19, Mac-1, Gr-1, Ter-119, CD3ε, CD5, and CD8α. BM ILC1 were defined as Lin−Thy1.2+IL-7R+CD62L−CD27+ST2−; BM ILC3 as Lin− Thy1.2+IL-7R+CD62L−CD27−ST2−; and BM NK as Lin−Thy1.2+IL-7R−CD62L+NK1.1+DX5+. Lineage markers for BM pre-cDC and BM pDCs are CD3, CD8α, CD5, TCRβ, γδTCR, NK1.1, Ter119, and Ly6G. BM pre-cDCs were gated as Lin−B220−MHCII−CD11c+Flt3+Sirpα− and BM pDCs as Lin−B220+CD19−SiglecH+. Thy NKs were considered as Lin (CD3, CD8α, CD4, TCRβ, ΥδTCR)−CD122+NK1.1+DX5+. Data shown are pooled from three independent experiments (n = 8–10). Student’s t test was used to determine statistical significance. Error bars are SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. (c) Thymocytes from mice used in panel a were cultured in the presence of PMA and ionomycin plus monensin for 4 h before intracellular staining and analyzed with indicated antibodies. Data shown are representatives from two independent experiments.