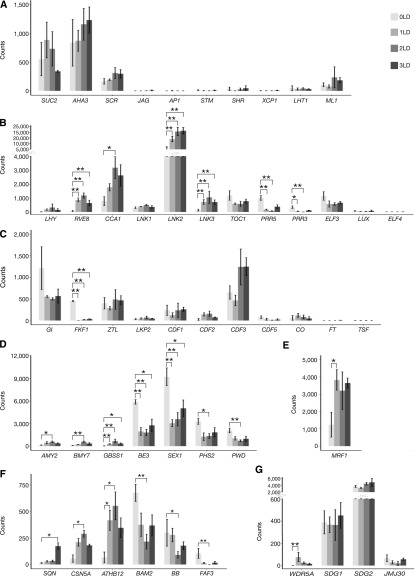
Figure 2.
Differential Gene Expression in the PCCs during LD Induction of Flowering.
Expression is given as count of sequencing reads. Gray gradient colors indicate the time points before (lighter gray) and 1 d (light gray), 2 d (dark gray), and 3 d (darker gray) after the shift to LD, respectively. *P < 0.05, **P < 0.01 (Deseq2 P values corrected for multiple testing using the Benjamini-Hochberg method).
(A) Expression of marker genes for the PCCs (SUC2 and AHA3), bundle sheath (SCR), leaf primordia (JAG), flower primordia (AP1), meristem (STM), xylem (SHR and XCP1), mesophyll cells (LHT1), and epidermis (ML1).
(B) Expression of marker genes for circadian clock, LHY, RVE8, CCA1, LNK1, LNK2, LNK3, TOC1, PRR5, PRR3, ELF3, LUX, and ELF4.
(C) Expression of marker genes for photoperiod flowering pathway, GI, FKF1, ZEITLUPE, LOV KELCH PROTEIN2, CDF1, CDF2, CDF3, CDF5, CO, FT, and TSF.
(D) Expression of important genes involved in carbohydrate metabolism, α-AMYLASE2, β-AMYLASE7, GBSS1, BE3, SEX1, PHS2, and PHOSPHOGLUCAN WATER DIKINASE.
(E) Expression of MRF1.
(F) Expression of selected important genes involved in development, SQUINT, CSN5A, ATHB12, BAM2, BB, and FAF3.
(G) Expression of selected epigenetic modifiers involved in regulation of circadian clock and photoperiodic flowering time, WDR5A, SDG1, SDG2, and JMJ30.