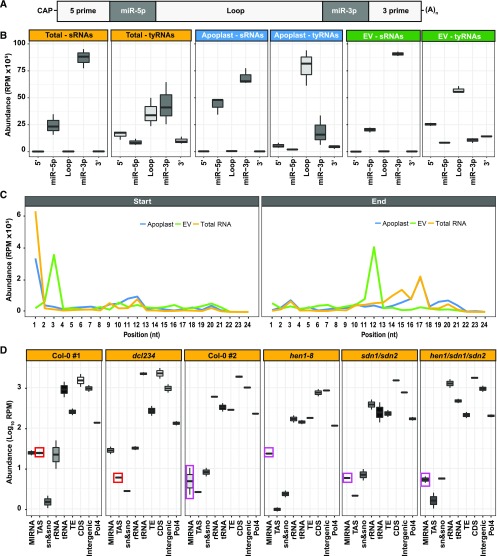
Figure 2.
EV tyRNAs Are Derived from Specific Regions of Precursor RNAs.
(A) miRNA precursor structure, including from left to right, 5′ region, mature miRNA miR-5p, loop region, mature miRNA miR-3p, and 3′ region.
(B) EV tyRNAs are enriched in sequences that map to the loop region of miRNA precursors. The abundance of sRNAs and tyRNAs mapping to different regions of a miRNA precursor, as described in (A). The y axis represents abundance, in reads per million (RPM).
(C) EV tyRNAs mapping to mature miRNAs are cut out of the central region. Plots show the relative position of 5′ ends (left plot) or 3′ ends (right plot) of tyRNAs relative to nucleotide position within the parent miRNA. RPM, reads per million.
(D) Mutations that affect abundance of specific subclasses of sRNAs have a corresponding influence on tyRNA abundance. The abundance of total leaf tyRNAs mapping to different features of the Arabidopsis genome. These include the following from left to right: miRNA precursors, TAS precursors, snRNA and snoRNAs; rRNAs; tRNAs; TEs; CDSs (including introns and UTRs); intergenic sequences; and known Pol4 precursors. The y axis represents abundance, in reads per million (RPM), in a logarithmic (Log10) scale. Colored boxes highlight tyRNAs affected by mutations.