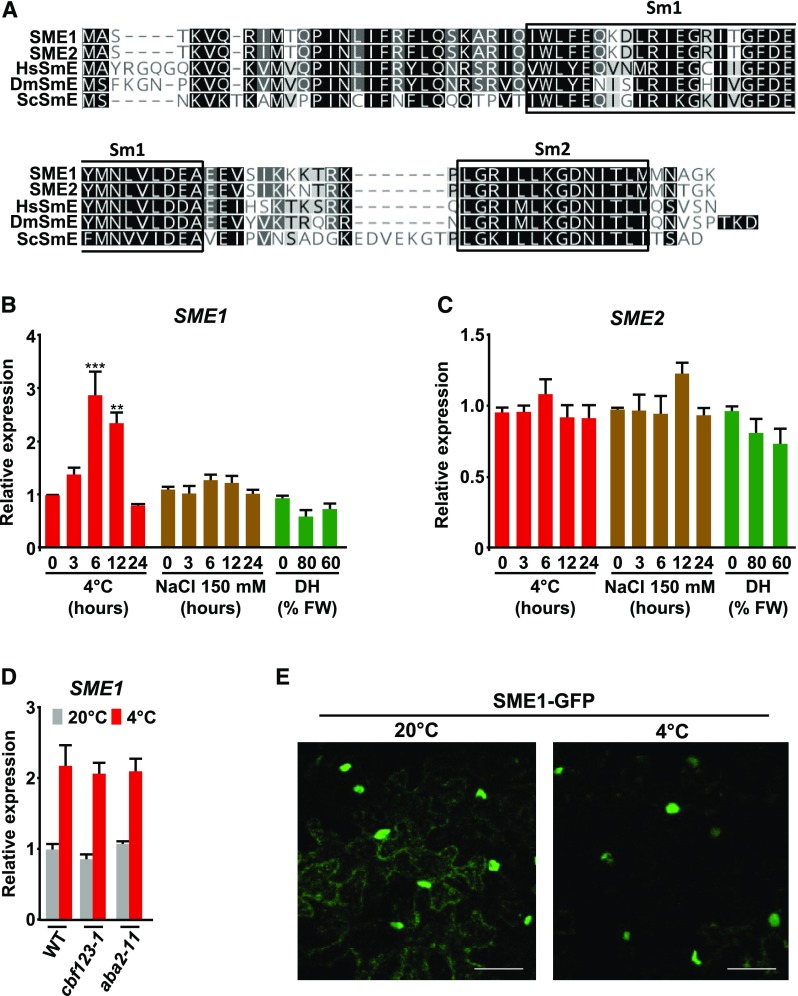
Figure 1.
Low Temperature Promotes SME1 Expression and Nuclear Localization of the Corresponding Protein.
(A) Sequence alignment of SME proteins from Arabidopsis, Homo sapiens, Saccharomyces cerevisiae, and Drosophila melanogaster. Sm1 and Sm2 domains are shown. Black and gray shading indicates identical or similar residues, respectively, in at least half of the compared sequences. Dashes represent gaps inserted into the sequences for optimal alignment.
(B) and (C) Accumulation of SME1 (B) and SME2 (C) transcripts in 2-week-old wild-type (WT) plants exposed for the indicated number of hours to 4°C (red bars) or 150 mM NaCl (brown bars), or dehydrated (DH) until reaching the indicated percentage of fresh weight (FW) (green bars).
(D) Accumulation of SME1 transcripts in 2-week-old wild-type, cbf123-1, and aba2-11 plants grown under control conditions (20°C; gray bars) or exposed for 6 h to 4°C (red bars).
(E) Subcellular localization of SME1-GFP in N. benthamiana leaf cells under control (20°C) or cold (4°C, 24 h) conditions. Scale bars indicate 50 µm.
In (B) to (D), transcript levels, determined by qPCR, are represented as relative to values at 0 h or 0% DH (B and C) or to wild-type values (C). Data represent the mean of three independent experiments. Asterisks indicate significant differences (**P < 0.001, ***P < 0.0001) between treated and control (0 h) plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.