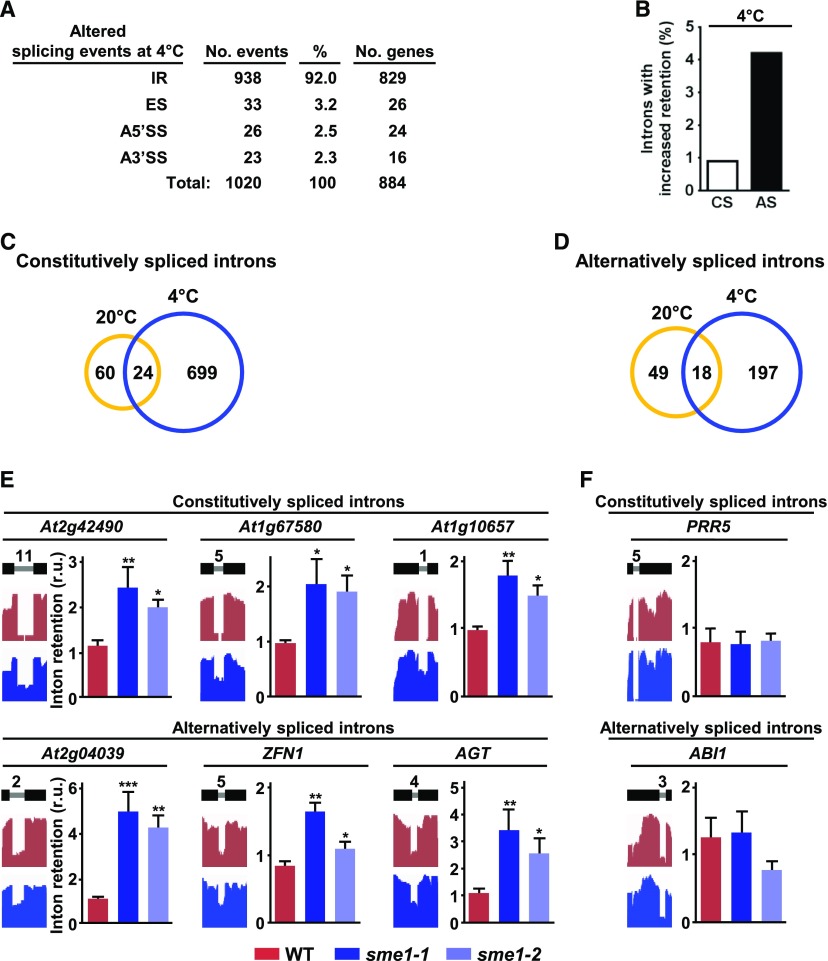
Figure 6.
Arabidopsis SME1 Controls the Splicing of Specific Constitutive and Alternative Spliced Introns in Response to Low Temperature.
(A) Quantification of altered splicing events (IR, ES, A5′SS, A3′SS) identified in sme1-1 with respect to wild-type (WT) plants in response to low temperature. The number of different genes affected in each case is also shown.
(B) Percentage of constitutively (CS) and alternatively (AS) spliced introns with increased retention in sme1-1 plants exposed to 4°C relative to the total number of the corresponding introns identified in wild-type plants exposed to the same conditions.
(C) and (D) Venn diagrams showing the overlap between the constitutively (C) and alternatively (D) spliced introns specifically retained in sme1-1 with respect to wild-type plants under control (20°C) or cold (4°C) conditions. The number of specific and common introns identified is indicated.
(E) Validation of different IR events identified in sme1-1 plants exposed to low temperature. In each case, the name of the gene containing the corresponding constitutively or alternatively spliced intron is indicated.
(F) Validation of constitutively and alternatively spliced introns not affected in sme1-1 plants exposed to low temperature. The names of the genes containing the corresponding introns are indicated.
In (E) and (F), a diagram of the pre-mRNA regions, including the considered introns (gray lines) with their relative positions in the representative gene model and the flanking exons (black bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software, is shown in the left. The quantification of considered introns in wild-type, sme1-1, and sme1-2 plants by qPCR assays is displayed in the right. Data represent the mean of three independent experiments. Asterisks indicate significant differences (*P < 0.01, **P < 0.001, ***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.