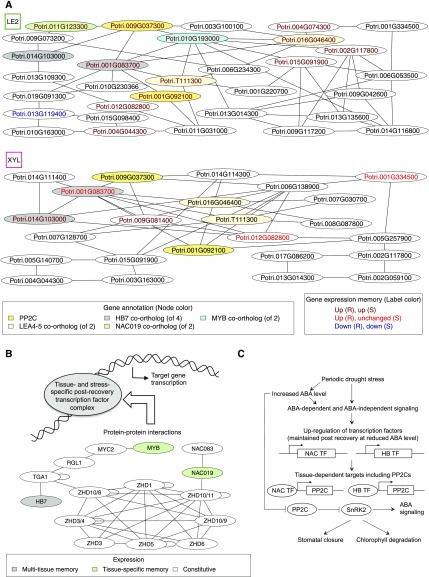
Figure 8.
Predicted Regulatory Stress-Related Memory Processes in Mature Leaves (LE2) and Developing Xylem (XYL).
(A) Core regulatory networks obtained by iteratively removing single-edge nodes from expression-based regulatory network predictions (Figure 7A). Node label colors refer to periodic stress-related expression patterns. Function annotation is shown for selected nodes discussed in the main text. Potri.010G193000 is a co-ortholog of the Arabidopsis thaliana MYB transcription factor AT5G05790, here abbreviated as MYB.
(B) Model of possible transcription factor complex formation in stress-related memory derived from protein-protein interaction data in Arabidopsis. Gene names are taken from the orthology information in Phytozome. For some ZHD ortholog groups, different Arabidopsis genes (marked by an Arabidopsis ZHD identifier after the slash) constitute the best BLASTP matches of poplar genes.
(C) Model suggesting physiological roles of PP2Cs and regulatory transcription factors in mature leaves during and after periodic stress.