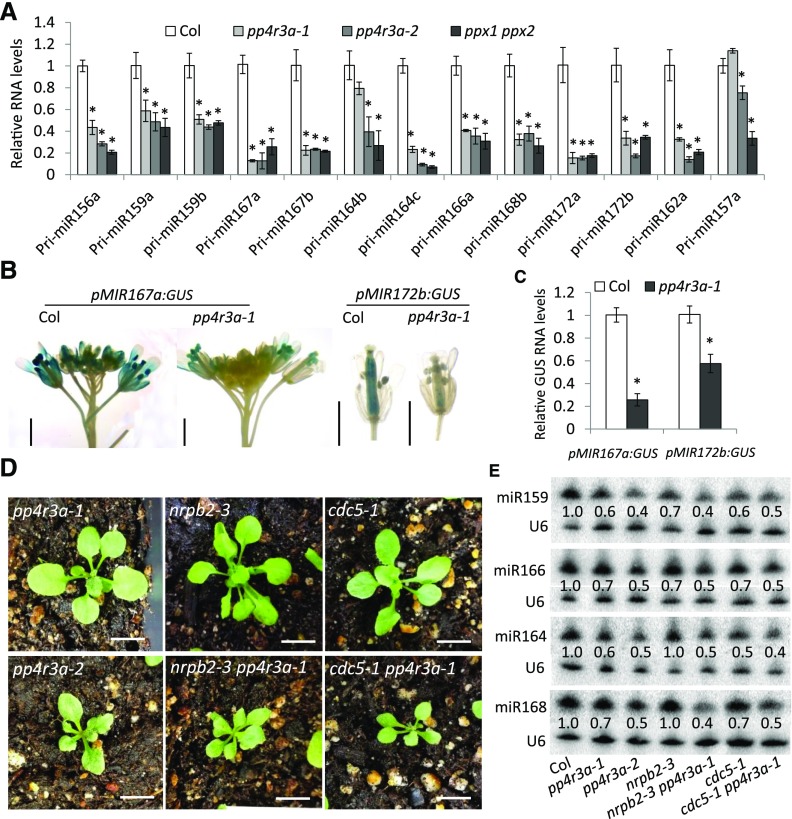
Figure 5.
Requirement of PP4R3A for MIR Transcription.
(A) Determination of pri-miRNA levels by RT-qPCR.
(B) GUS staining of representative samples harboring pMIR167a:GUS and pMIR172b:GUS in the Col and pp4r3a-1 backgrounds. Scale bars = 2 mm.
(C) GUS transcript levels in samples harboring pMIR167a:GUS and pMIR172b:GUS in the Col and pp4r3a-1 backgrounds, as determined by RT-qPCR.
(D) and (E) Genetic interactions between mutations in PP4R3A, NRPB2 and CDC5. (D) Additive phenotypes of nrpb2-3 pp4r3a-1 and cdc5-2 pp4r3a-1 double mutants. Scale bars = 1 cm. Note that the phenotypes of the double mutants were much stronger than the corresponding single mutants, but similar to that of pp4r3a-2. (E) The abundance of miRNAs in Col, pp4r3a-1, pp4r3a-2, nrpb2-3, cdc5-1, nrpb2-3 pp4r3a-1, and cdc5-1 pp4r3a-1, as detected by RNA gel blot analysis of small RNAs.
In (A) and (C), UBQ5 served as the internal control; error bars indicate sd from three independent experiments; asterisks indicate significant difference (t test, P < 0.05).