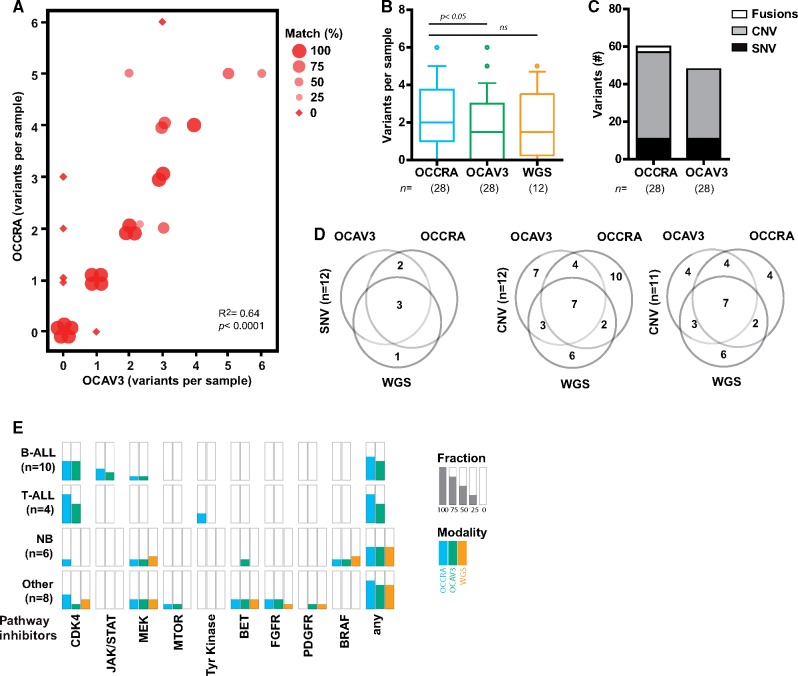
Figure 1.
Improved variant discovery using childhood cancer-specific amplicon-based sequencing. A) Variant discovery across 28 tumor samples using Oncomine Childhood Cancer Research Assay (OCCRA) and Oncomine Comprehensive Assay Version 3 (OCAV3) next-generation sequencing assays. The size and shape of the marker indicates the percentage of concordance (identical variants/total variants) between the assays. B) OCCRA detected more variants per sample than OCAV3 (n = 28, paired t-test, two tail). For the 12 samples analyzed by three modalities, the variants detected per sample were not statistically different (n = 12, one-way ANOVA with Tukey multiple comparison). Data plotted as box and whiskers (10–90 percentile). C) The types of variant detected included more copy number variants (CNV) and fusions in samples assayed by OCCRA than parallel analyses with OCAV3. D) Venn diagrams for variants in pediatric cancer driver genes detected by each assay compared to whole genome sequencing (WGS) shows high concordance in the detection of single nucleotide variants (SNV) but less for CNVs, which is improved by the removal of sample No. 18 (20% tumor content). E) The fraction of samples that were matched with the indicated pathway inhibitors for each modality across the indicated tumor types.