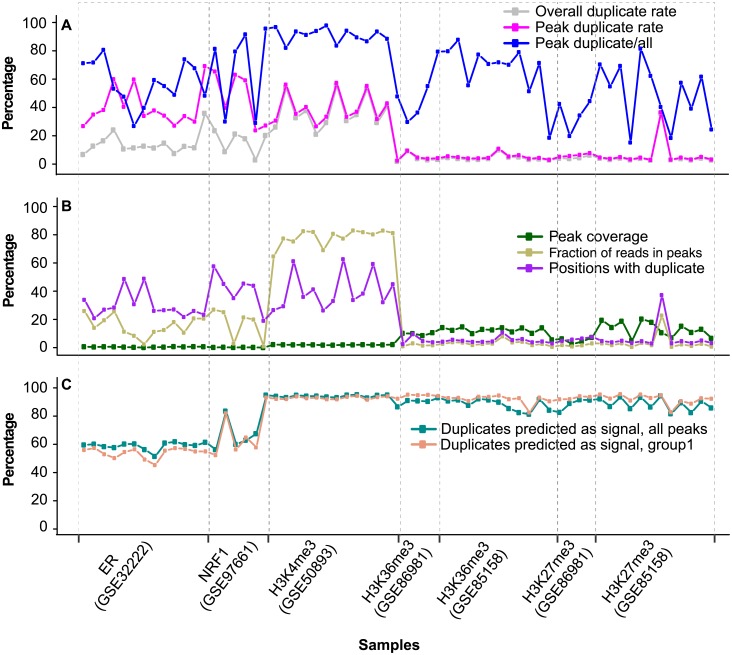
Fig 6. Prediction of duplicates as signal based on peak enrichment.
(A) Duplicate rate in a library and in peaks and proportion of duplicates in peaks. Duplicate rate in a lib was estimated as the number of duplicates divided by the number of uniquely mapped reads. Duplicate rate in peaks was estimated in the same way. (B) Plot of peak coverage, fraction of positions with duplicates and fraction of nonredundant reads in peaks. Peak coverage was estimated as the total peak size over the mappable genome size (0.75 x genome size). Fraction of positions with duplicates was estimated as the number of positions with duplicates over the number of positions with uniquely mapped reads. Fraction of reads in peaks (FRiP), fraction of uniquely-mapped, nonredundant reads in peaks. (C) Proportion of duplicates predicted as signal. The prediction was based on the correlation between peak duplicate and non-duplicate level, as showed in Fig 4.