Fig. 1. Ultra-multiplexed, automated cell culture system for dynamical live-cell analysis.
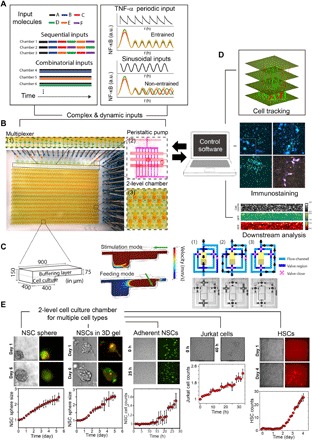
The microfluidic device contains 1500 independently programmable culture chambers. During a 1-week experiment, the device performs nearly 106 pipetting steps to create and maintain distinct culture conditions in each of the chambers. (A) Each chamber can execute a distinct dynamic culture program (combinations, timed sequences, sine waves, etc.) where the fluidic composition can be changed when desired, and dynamic processes (i.e., NF-κB localization or Hes5 expression) are tracked with single-cell resolution. An on-chip nanoliter multiplexer measures several fluids and mixes them at predetermined ratios to create complex chemical inputs. A peristaltic pump delivers inputs to any given chamber. For the combinatorial input scenario, several chemicals are mixed and delivered to the cells continuously. In sequential inputs, signaling molecules are changed with a programmed time interval (Δt = 1 day). a.u., arbitrary units. (B) The system can culture adherent or nonadherent cells in either suspension mode, monolayer populations, or 3D format using hydrogels. The novel two-layer geometry of the culture chambers allows diffusion-based media delivery to create a stable environment for cells, and provides the additional ability of single-cell tracking of even nonadherent cells during dynamical stimulation. (C) Left: Two-layer cell chamber design allows diffusion- or flow-based media delivery, 3D cell culture, immobilization of nonadherent cells by gravity, and automated cell retrieval. Middle: Fluid mechanical simulations indicate the flow rates for diffusion-based media delivery and cell retrieval via direct flow. Right: Each chamber is controlled by a network of dedicated channels and membrane valves that automate various cell culture procedures. (D) Cells can be immunostained in the chip. The system is integrated to a fluorescent microscope and can automatically track individual cells in time-lapse experiments. Single cells or populations of interest can be automatically retrieved from individual chambers for off-chip analysis or expansion. GFP, green fluorescent protein; RFP, red fluorescent protein. (E) Primary cells (e.g., mouse NSCs and human HSCs) and cell lines (e.g., Jurkat T cells and mouse fibroblasts) are viably cultured and maintained on chip for weeks. Growth rates equal or better than the well plate culture are achieved through frequent diffusion-based media delivery while maintaining an unperturbed microenvironment.
