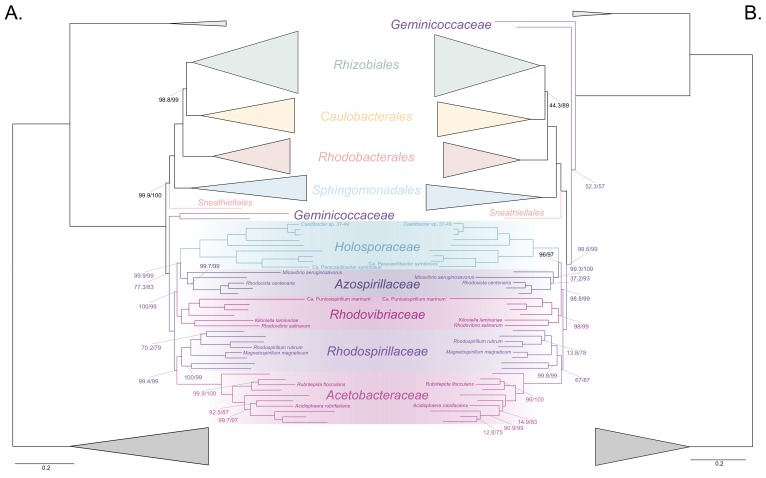
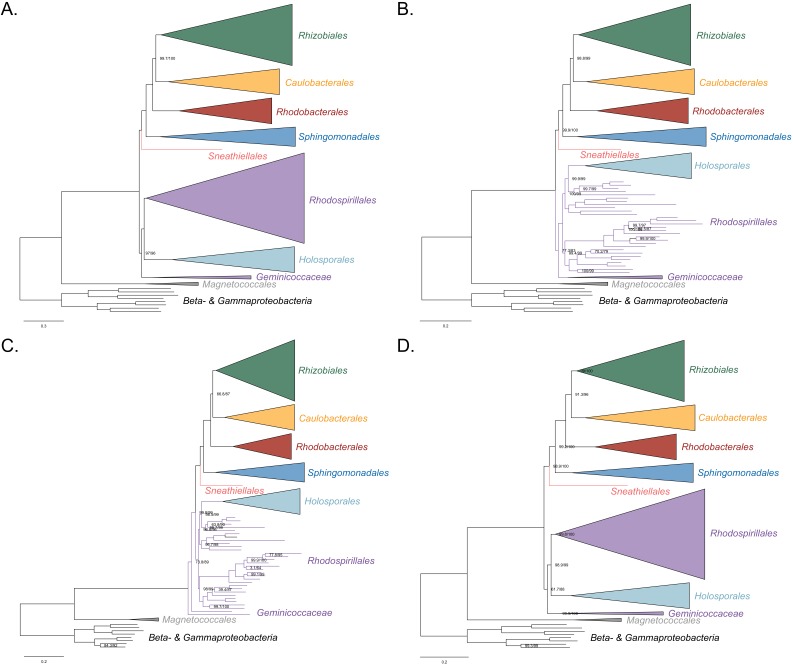
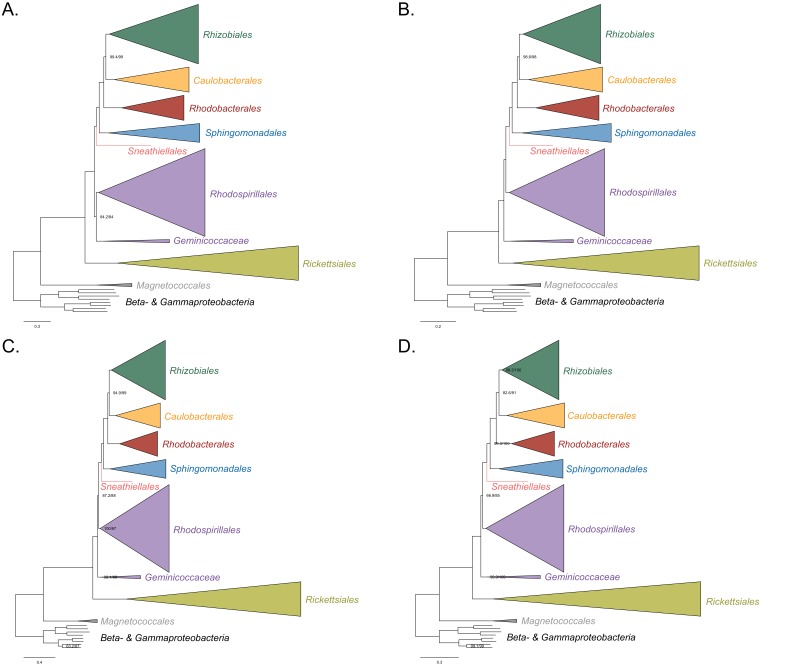
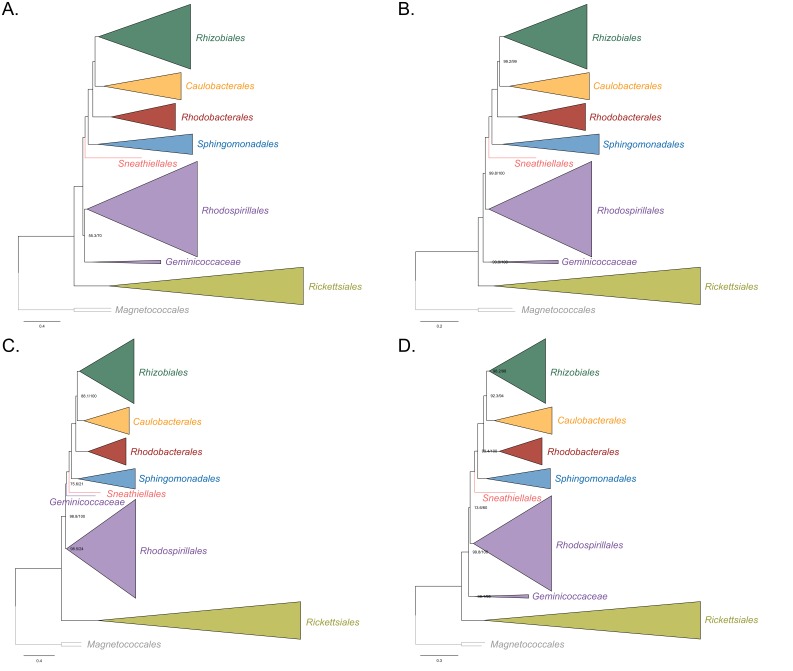
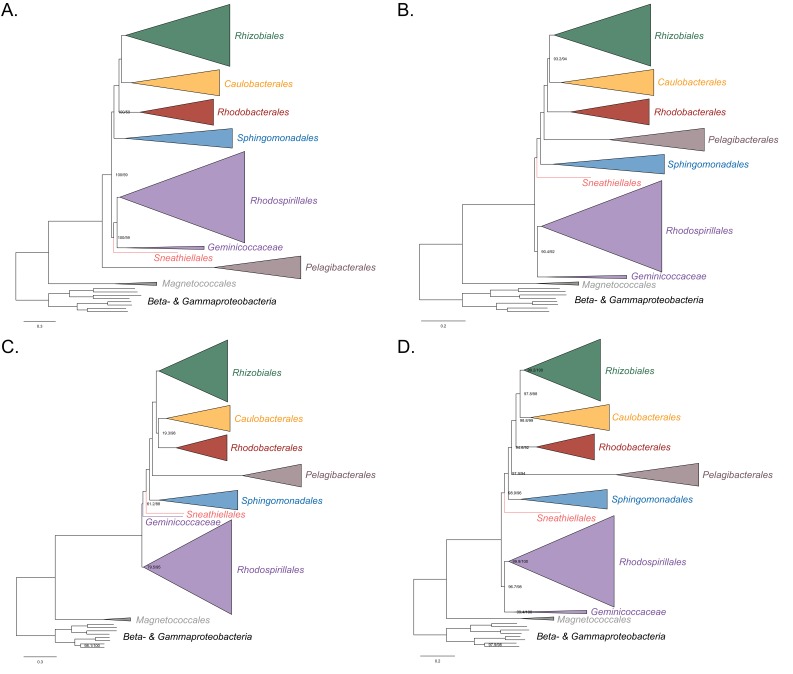
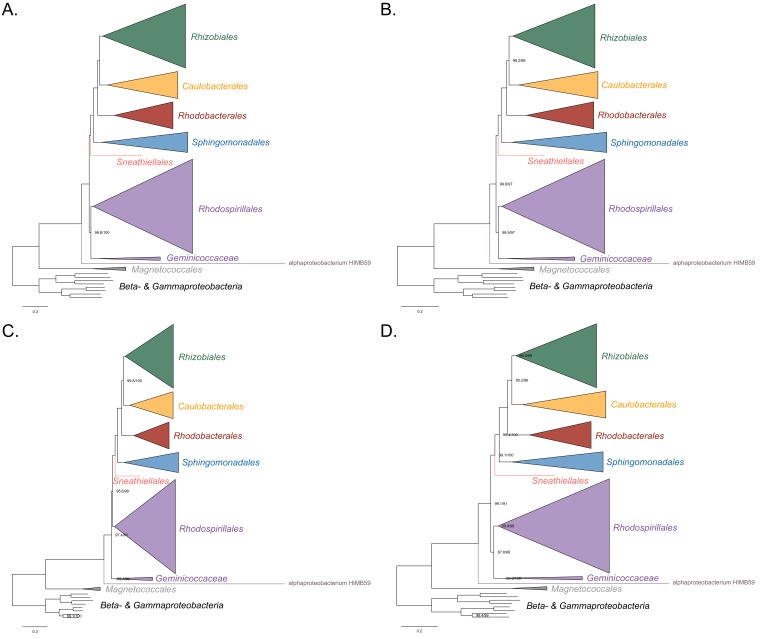
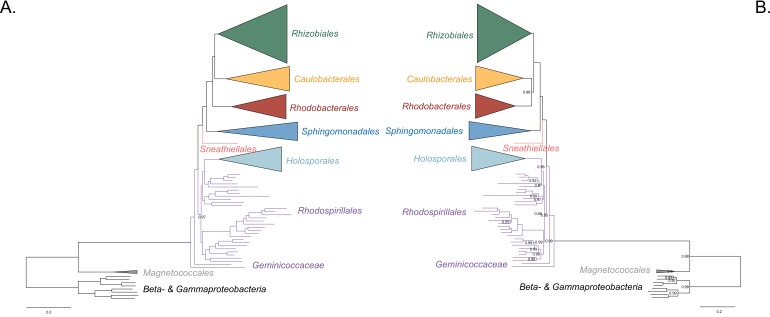
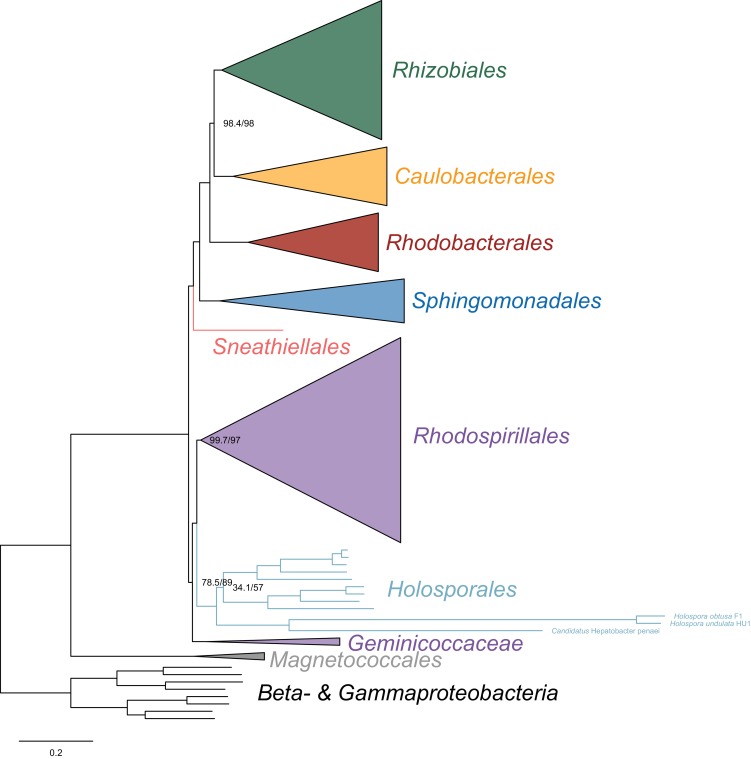
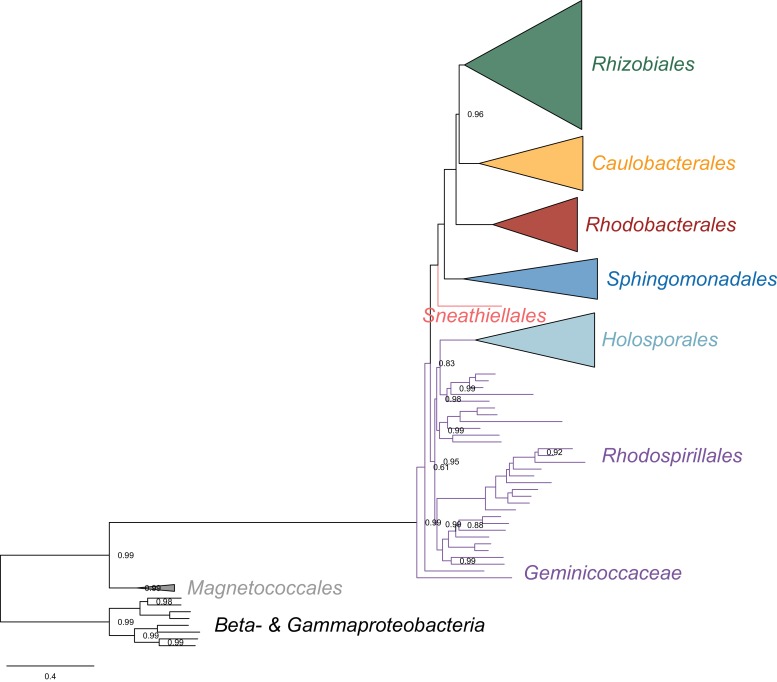
Figure 3. The Holosporales (renamed and lowered in rank to the Holosporaceae family here) branches in a derived position within the Rhodospirillales when compositional heterogeneity is reduced and the long-branched and compositionally biased Rickettsiales, Pelagibacterales, and alphaproteobacterium HIMB59 are removed.
Branch support values are 100% SH-aLRT and 100% UFBoot unless annotated. (A) A maximum-likelihood tree, inferred under the LG + PMSF(ES60)+F + R6 model, to place the Holosporaceae in the absence of the Rickettsiales, Pelagibacterales, and alphaproteobacterium HIMB59 and when compositional heterogeneity has been decreased by removing 50% of the most biased sites. The Holosporaceae is sister to the Azospirillaceae fam. nov. within the Rhodospirillales. (B) A maximum-likelihood tree, inferred under the GTR + ES60 S4+F + R6 model, to place the Holosporaceae in the absence of the Rickettsiales, Pelagibacterales, and alphaproteobacterium HIMB59, and when the data have been recoded into a four-character state alphabet (the dataset-specific recoding scheme S4: ARNDQEILKSTV GHY CMFP W) to reduce compositional heterogeneity. This phylogeny shows a pattern that matches that inferred when compositional heterogeneity has been alleviated through site removal. See Figure 3—figure supplement 6 for the Bayesian consensus trees inferred in PhyloBayes MPI v1.7 and under the and the CAT-Poisson+Γ4 model. See also Figure 3—figure supplements 1–5 and 7–8.