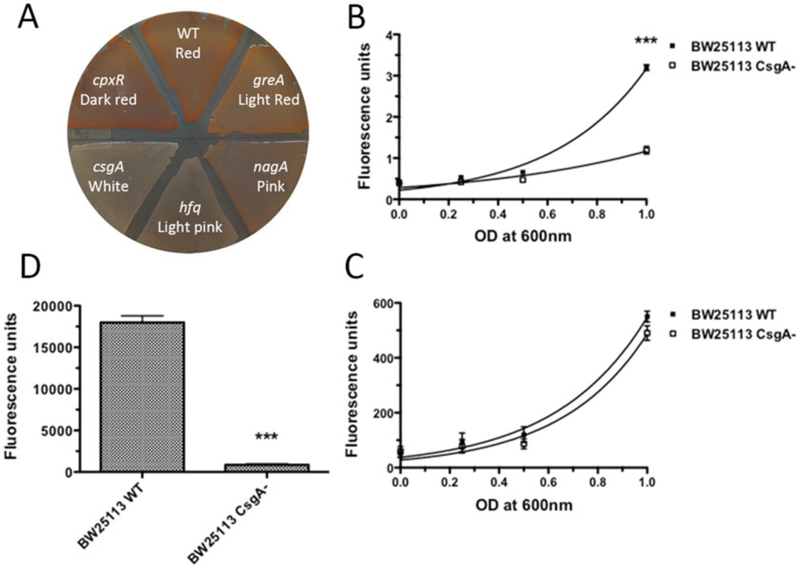
Fig. 1.
CR staining and fluorescence quantification. (a) Use of CR staining to screen for curli mutants. Colonies of the wide type E. coli stained red on a YESCA CR agar plate while the curli defective mutant, csgA, remained white. Curli mutants [56] showed dark red, pink or light pink color on the YESCA CR plate. (b–d) Measurements of the fluorescence associated with bacteria for CR (Em/Ex: 485/612 nm). BW25113 wild type and the csgA deletion mutant strains were recovered from YESCA plates after 48 h of growth at 26 °C and resuspended in 1 mL of 50 mM KPi (pH 7.2) containing 0.5 μg/mL CR or 4.5 μg/mL DAPI. After wash, serial base two dilutions are prepared and the fluorescence was measured in a 96-well plates by triplicate. As a reference, 100 μL of 50 mM KPi were used. (b) Comparison of CR fluorescence of curli producing wild type and curli deficient csgA strains. Non-linear (exponential fit): R2 = 0.970 and 0.778 for BW25113 and csgA, respectively. (c) To show that the BW25113 csgA mutant was present in the same amount as the wild type BW25113 strain, bacteria were post-labeled with the unspecific DNA 4′,6-diamidino-2-phenylindole (DAPI) fluorochrome and the DAPI fluorescence is measured (Em/Ex: 350/460 nm). Non-linear (exponential fit): R2 = 0.865 and 0.867 for BW25113 and csgA, respectively. (d) Quantification of CR fluorescence for bacteria pre-stained on YESCA CR plates. The CR fluorescence of the wild type strain was significantly higher (around three times) compared to the BW25113 csgA mutant. ***: p < 0.0001 using student’s t test. Error bars: average SEM of at least 6 wells per sample. The plate reader used in this determination was an Infinite 200 with the Tecan-I application and automatic optimization of the gain