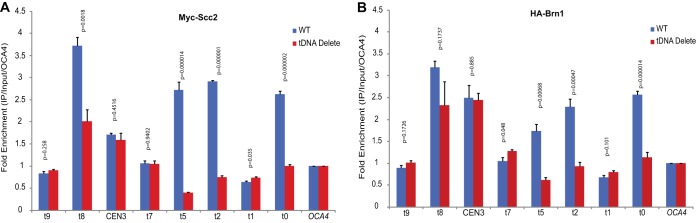
FIG 4.
Scc2 and Brn1 binding at tDNAs on chromosome III. (A) ChIP-qPCR mapping of Myc-Scc2. The data show the distribution of Scc2 at specific sites along chromosome III in the wild-type and tDNA delete strains. The data are the results of two independent cross-links on which four IPs were performed. For each amplicon, the fold enrichment compared to input was first calculated, and the data were then normalized to the OCA4 locus. An unpaired t test assuming unequal standard deviations (SD) was used to test for significance of differences between the wild-type and tDNA delete strains. (B) ChIP-qPCR mapping of HA-Brn1 and condensin. Fold enrichment and statistical significance were calculated in the same way as for the Scc2 ChIP and normalized to the OCA4 locus. The error bars indicate SD.