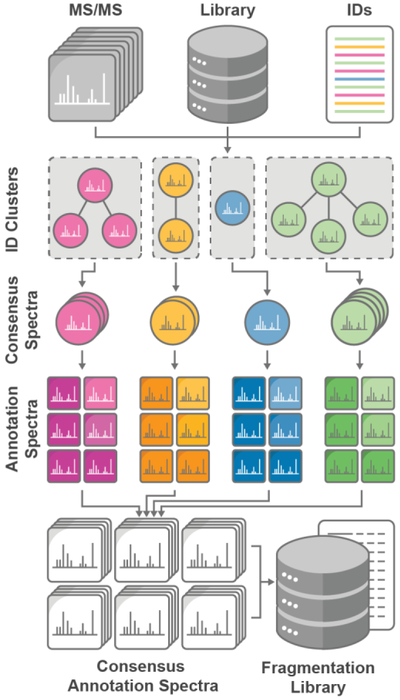
Figure 1.
Spectral Library Generation Workflow. Lipid fragmentation mapping requires raw MS/MS spectra, a library of potential lipid class metadata, and putative identifications (complex method) or standard retention times (reference standard method). Duplicate identifications are clustered and curated to generate consensus MS/MS spectra. Annotation spectra are subsequently generated from transformed MS/MS peaks and combined to reveal the fragment definitions and intensities common to a lipid class. Derived fragmentation rules are then used to generate a new tailored in-silico spectral library.