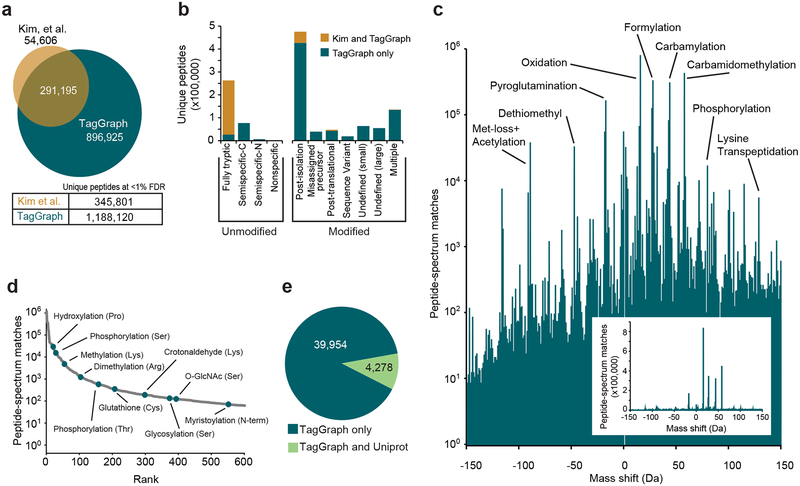
Figure 3. TagGraph extends deep proteome characterization to post-translational modifications.
a) TagGraph confirmed the majority of identifications made by Kim et al.24 (Dataset 5 (https://taggraph.page.link/Datasets)), but also expanded unique peptide identities from the human proteome dataset over three-fold relative to those originally reported. b) Categorical breakdown of unique peptide forms (distinguishing PTMs) identified by TagGraph. As expected, the majority of peptides identified by both TagGraph and Kim et al. correspond to tryptic peptides. Peptides identified by TagGraph but not Kim et al. primarily originated from non-tryptic peptides and peptides with unanticipated modifications. Post-isolation modifications comprised the most prevalent identification category in this dataset. c) Mass shifts (modified amino acid mass – unmodified amino acid mass) corresponding to all modifications identified by TagGraph from the human proteome dataset reveal a complex modification landscape. Numbers of identifications (peptide-spectrum matches) span six orders of magnitude. Despite the presence of several highly abundant post-isolation modifications (e.g., formylation), the depth of the proteomic profiling achieved in this dataset made it possible to characterize lower abundance post-translational modifications. Inset: modification frequencies without log transformation. d) Ranked relative abundances of 2,576 PTM-amino acid combinations, as estimated by the number of spectra bearing each from the human proteome dataset. Ten of these are highlighted; all modifications are represented in Dataset 9 (https://taggraph.page.link/Datasets). e) Over 90% of modification sites (39,954 of 44,232) identified by TagGraph from the human proteome dataset were not among 52,959 previously described in Uniprot. However, the overlap in the sites reported by TagGraph and Uniprot (4,278) is highly significant (p-value = 1e-308, one-tail Fisher’s exact test).