FIGURE 3.
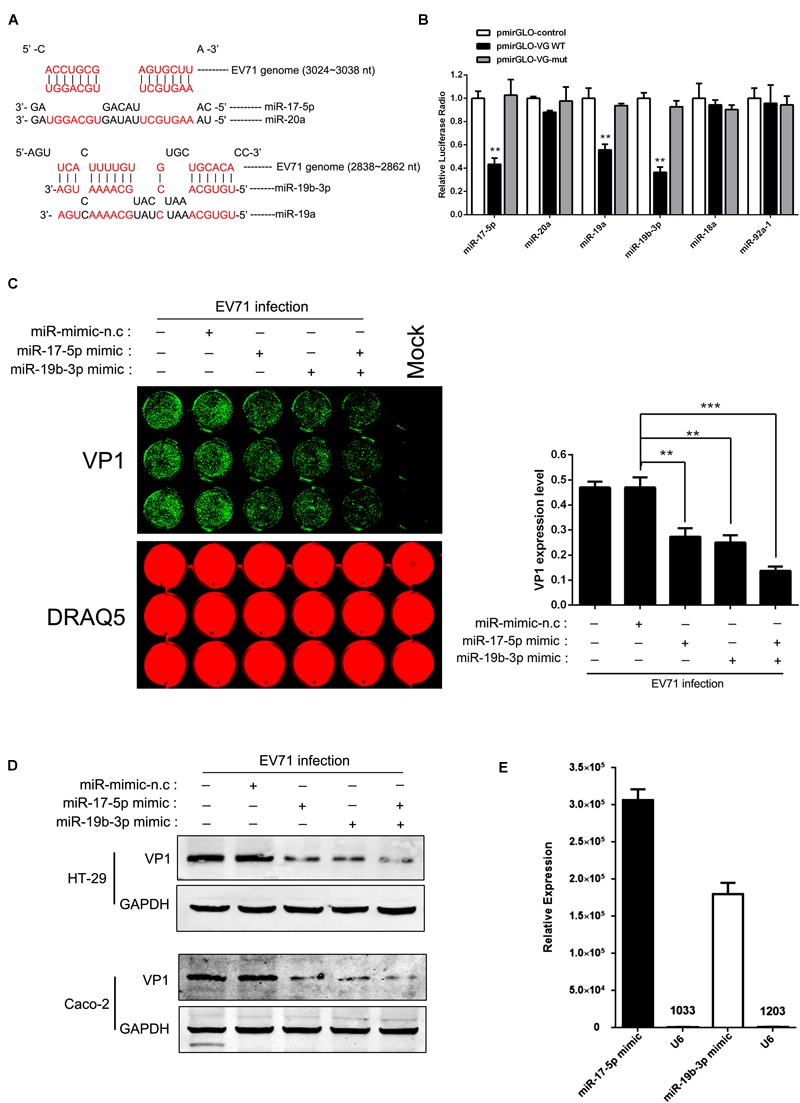
miR-17-5p, miR-19a, and miR-19b directly targeted the EV71 genome. (A) Potential target sites of mature miRNAs of the miR-17-92 cluster in the EV71 genome. Computational analysis with RNA-hybrid software yielded potential binding sites for miR-17, miR-20a, and miR-19a/b in the EV71 genome. (B) EV71 genome containing presumptive miR-17-92 target sequences, was cloned into the multiple cloning sites at the 3′ of the firefly luciferase reporter gene (luc2) in the luciferase expression plasmid (pmirGLO). Either control plasmid (pmirGLO-control), pmirGLO-viral genome WT (pmirGLO-VG WT) or pmirGLO-viral genome mut (pmirGLO-VG-m) was co-transfected with individual miRNA mimic in HT-29 cells. The luciferase activity of pmirGLO-VG WT group was repressed by transfecting miR-17 and miR-19a/b while mutation of a predicted miRNAs binding sites abolished miRNAs-mediated repression. (C,D) HT-29 cells and Caco-2 were infected with EV71 (MOI = 1) after being transfected with miR-17 or miR-19b mimic for 24 h. EV71 VP1 expression level was determined via In-cell Western and normalized by DRAQ5 (C) or Western blot (D). The symbols “+” and “-” indicate treatment and no treatment, respectively, by the corresponding factor. (E) HT-29 cells transfected with miR-17 or miR-19b mimic were subjected to Ago2-IP, and copy numbers of miR-17 and miR-19b in the immunoprecipitates were determined by real-time quantitative PCR, using the U6 as a negative control. All the results shown were representative of three independent experiments and data were presented as mean ± SEM (∗∗P < 0.01, ∗∗∗P < 0.001).
