Figure 4.
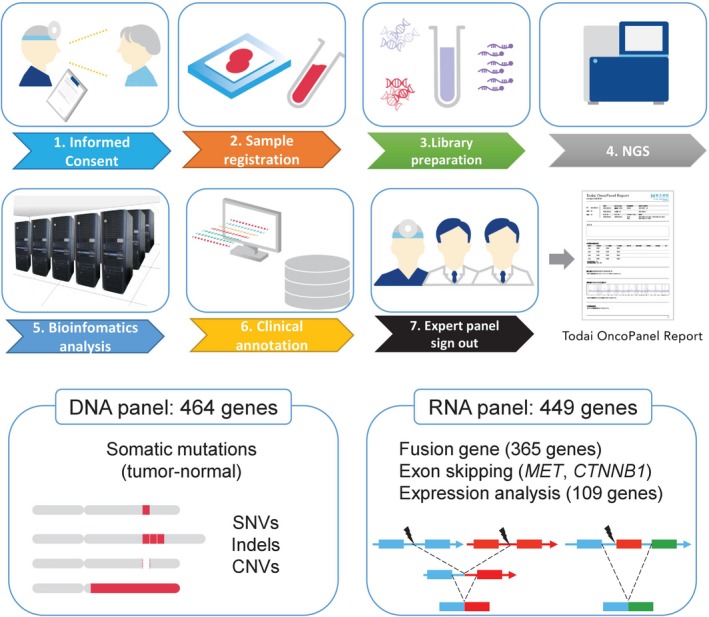
Overview of the TOP workflow. Patients provide informed consent for paired tumor‐normal sequence analysis, and a blood sample is collected as a source of normal DNA. DNA is extracted from the tumor and blood samples, and sequence libraries are prepared and captured using hybridization probes targeting all coding exons of 463 genes and the TERT promoter region. RNA is extracted from tumor samples and reverse transcribed. cDNA sequence libraries are prepared and captured using hybridization probes targeting 365 fusion genes, MET and CTNNB1 exon skipping, and 109 genes for expression analysis. Following sequencing, paired reads are analyzed through a custom bioinformatics pipeline that detects multiple classes of genomic and transcriptional alterations. The results are loaded into a genomic variants database developed in house, where they are manually reviewed for quality and accuracy. After expert panel (molecular tumor board) review of each case, the final version of each TOP report is signed out and transmitted to the electronic medical record. CNV, copy number variant; NGS, next‐generation sequencing; SNV, single nucleotide variant
