Figure 5.
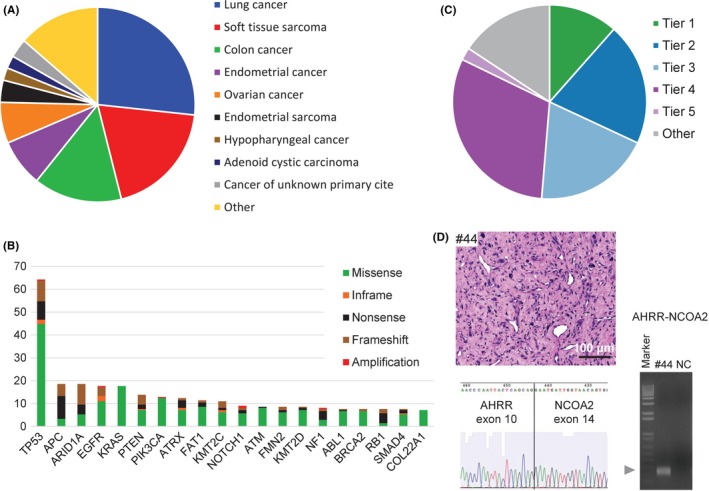
Overview of the TOP prospective cohort. A, Distribution of tumor types among the cases successfully sequenced from 183 patients. Cases represented more than 10 principal tumor types. B, The 20 most recurrent somatic alterations. Bars indicate the percentage of cases harboring the mutations, and the types of genomic alterations are color‐coded. C, Clinical actionability of somatic alterations revealed by the TOP. Alterations were annotated based on their clinical actionability according to TOP classification (Figure S10), and samples were assigned to the level with the most actionable alteration. Briefly, the levels of evidence vary according to whether the mutations are a Pharmaceuticals and Medical Devices Agency‐recognized biomarker (Tier 1), a Japanese clinical trial‐targeted biomarker or a US Food and Drug Administration‐recognized biomarker (Tier 2), an investigational agent sensitizing gene alteration or an oncogenic alteration supported by a knowledge database (Tier 3), an oncogenic alteration supported by a research paper (Tier 4) or a recurrently reported alteration in a knowledge database. D, A representative photograph of tumor #44 stained with hematoxylin‐eosin (scale bar, 100 μm; upper left panel). The proliferation of spindle cells with atypical nuclei and surrounding myxoid interstitial tissue were observed. A fresh frozen sample from case #44 was subjected to reverse transcription (RT) polymerase chain reaction with the fusion‐RT primer set to detect AHRR‐NCOA mRNA (lower left panel). The arrowhead indicates the estimated sizes of the fusion transcript. The band was extracted from the gel and subjected to Sanger sequencing. The electrophoretogram obtained from the band supported the junction sequences of AHRR‐NCOA2 (right panel). Marker, 1‐kb DNA ladder; NC, negative control
