Abstract
Recent studies have identified several microRNAs (miRNAs/miRs) that are dysregulated in clear cell renal cell carcinoma (ccRCC), and their dysregulation may serve important roles in the occurrence and development of ccRCC. Therefore, understanding the expression pattern and functional roles of miRNAs in ccRCC may facilitate the identification of novel therapeutic targets for the treatment of ccRCC. In the current study, reverse transcription-quantitative polymerase chain reaction was used to determine miR-508 expression levels in ccRCC tissue samples and cell lines. The cell counting kit-8 and in vitro Transwell invasion assays were used to examine the effects of miR-508 overexpression on ccRCC cell proliferation and invasion, respectively. In addition, bioinformatics analysis and dual-luciferase reporter gene assays were used to investigate the underlying mechanism of miR-508 in ccRCC cells. Furthermore, the regulatory role of miR-508 on zinc finger E-box-binding homeobox 1 (ZEB1) mRNA and protein expression in ccRCC cells was investigated using RT-qPCR and western blot analysis, respectively. Additionally, the association between miR-508 and ZEB1 expression in ccRCC tissue samples was examined. Rescue experiments were performed to determine whether the tumor suppressive effects of miR-508 may be mediated by ZEB1 in ccRCC cells. The results of the current study demonstrated that miR-508 expression was significantly downregulated in ccRCC tissue samples and cell lines. In addition, miR-508 overexpression significantly decreased the proliferation and invasion of ccRCC cells. ZEB1 was identified as a direct target gene of miR-508 in ccRCC cells and the relative expression level of ZEB1 mRNA was significantly increased in ccRCC tissue samples. Furthermore, a negative correlation between miR-508 and ZEB1 expression was identified in ccRCC tissues. ZEB1 knockdown exhibited a functional role similar to miR-508 overexpression in ccRCC cells, and restoration of ZEB1 expression significantly reversed the inhibitory effects of miR-508 on the malignant phenotype of ccRCC cells. Taken together, the results of the current study demonstrated that miR-508 may serve a tumor suppressive role in ccRCC via direct targeting of ZEB1. MiR-508 may present a novel and efficient therapeutic target for the treatment of patients with ccRCC.
Keywords: clear cell renal cell carcinoma, microRNA-508, proliferation, invasion, zinc finger E-box-binding homeobox 1
Introduction
Renal cell carcinoma (RCC), originates from cells in the renal cortex and is the most common type of kidney cancer in adults worldwide, with ~270,000 new cases and ~116,000 RCC-associated mortalities each year (1,2). RCC is divided into three major subtypes, including clear cell RCC (ccRCC), papillary RCC and chromophobe RCC (3). ccRCC is the most common and aggressive of the three subtypes, and accounts for ~80–90% of all RCC cases (4). Despite recent advances in cancer treatment, the long-term prognosis of patients with advanced ccRCC is poor, with a median survival rate of 1.5 years (5). Metastasis, lymph node recurrence and cancer recurrence are the main factors associated with poor prognosis in patients with ccRCC (6). In ~33% of cases, patients with ccRCC are diagnosed with local or distant metastases due to its variable clinical presentation (7). In addition, there is a 40% recurrence rate in patients with ccRCC following surgery (8,9). Therefore, identifying novel therapeutic targets may improve the survival outcome and treatment response of patients with advanced ccRCC.
MicroRNAs (miRNAs/miRs) are a group of endogenous, short, non-coding RNAs ~18–23 nucleotides in length (10). MiRNAs regulate multiple biological and pathological processes, including carcinogenesis and cancer progression (11). miRNAs promote mRNA degradation or inhibition of mRNA translation by binding to the complementary binding sites in the 3′-untranslated region (3′-UTR) of target genes (12). Several studies have demonstrated that miRNA deregulation is common in almost all types of human cancer (13–15). Previous studies have identified several deregulated miRNAs associated with ccRCC, whereby oncogenic miRNAs are upregulated and tumor suppressor miRNAs are downregulated (16–18). Aberrantly expressed miRNAs are involved in the pathogenesis of ccRCC and are involved in numerous processes, including cell proliferation, cell cycle progression, apoptosis, metastasis, epithelial-to-mesenchymal transition and chemoresistance (19–21). miRNAs may therefore be used as potential biomarkers to improve early diagnosis and prognosis of patients with ccRCC.
Previous studies have demonstrated that miR-508 is abnormally expressed in several types of human cancer, including ovarian (22) and colorectal cancer (23) and esophageal squamous cell carcinoma (24). An additional study revealed that miR-508 is downregulated in RCC and may be involved in regulating cell migration and apoptosis (25). Therefore, the aim of the current study was to investigate the expression pattern and biological function of miR-508 in the pathogenesis of ccRCC, as well as investigate the underlying molecular mechanisms of miR-508 in the development and progression of ccRCC.
Materials and methods
Human tissue samples
A total of 21 ccRCC tissue and paired adjacent normal tissue samples were collected from patients that had undergone surgical resection at the Lanzhou University Second Hospital (Lanzhou, China) between May 2015 and February 2017. None of the patients had received radiotherapy or chemotherapy treatment prior to surgical resection. All tissue samples were taken immediately after tumor resection, frozen in liquid nitrogen and stored at −80°C. The current study was approved by the Ethics Committee of Lanzhou University Second Hospital and written informed consent was obtained from each patient. The clinicopathological features of patients with ccRCC are summarized in Table I.
Table I.
Clinicopathological characteristics of patients with ccRCC.
| Patient no. | Gender | Age (years) | TNM stage |
|---|---|---|---|
| 1 | M | 47 | T1aN0M0 |
| 2 | M | 51 | T2bN0M0 |
| 3 | M | 44 | T1bN0M0 |
| 4 | M | 26 | T3aN1M0 |
| 5 | F | 58 | T2bN0M0 |
| 6 | M | 63 | T2bN0M0 |
| 7 | M | 68 | T1aN0M0 |
| 8 | F | 53 | T2aN0M0 |
| 9 | F | 57 | T1aN0M0 |
| 10 | M | 64 | T1aN0M0 |
| 11 | M | 49 | T2bN0M0 |
| 12 | M | 63 | T1aN0M0 |
| 13 | M | 56 | T1aN0M0 |
| 14 | F | 55 | T3aN1M0 |
| 15 | M | 72 | T2bN0M0 |
| 16 | F | 64 | T2aN0M0 |
| 17 | M | 68 | T2bN0M0 |
| 18 | M | 53 | T2bN0M0 |
| 19 | F | 47 | T1bN0M0 |
| 20 | F | 55 | T2aN0M0 |
| 21 | M | 60 | T2aN0M0 |
ccRCC, clear cell renal cell carcinoma; TNM, tumor, node and metastasis; M, male; F, female.
Cell culture
The normal human renal cell line HK-2, the papillary RCC cell lines Caki-2 and ACHN, and two ccRCC cell lines 786-O and A498 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin (all purchased from Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and maintained at 37°C in a 5% CO2-humidified incubator.
Cell transfection
miR-508 mimics, negative control miRNA mimics (miR-NC), small interfering RNA (siRNA) targeting the expression of zinc finger E-box-binding homeobox 1 (ZEB1 siRNA) and negative control siRNA (NC siRNA) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The miR-508 mimics sequence was 5′-UGAUUGUAGCCUUUUGGAGUAGA-3′ and the miR-NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The ZEB1 siRNA sequence was 5′-GUCGCUACAAACAGUUGUATT-3′ and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The ZEB1-overexpression plasmid, pcDNA3.1-ZEB1 and empty pcDNA3.1 plasmid were synthesized by GeneCopoeia, Inc., (Rockville, MD, USA). Cells were seeded into six-well plates at a density of 6×105 cells/well. miR-508 mimics (100 pmol), miR-NC (100 pmol), ZEB1 siRNA (100 pmol), NC siRNA (100 pmol), pcDNA3.1-ZEB1 (4 µg) or empty pcDNA3.1 plasmids (4 µg) were transfected into cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. At 6 h following incubation at 37°C in a 5% CO2-humidified incubator, the medium was replaced with complete culture medium. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and cell proliferation assay was performed at 24 h post-transfection. Transwell invasion assay was performed in transfected cells following 48 h of incubation. Following 72 h culture, western blot analysis was utilized for the measurement of protein expression.
RT-qPCR
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to on the manufacturer's protocol. To examine the expression of miR-508, total miRNA was first reverse transcribed into cDNA using the TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The temperature protocol for reverse transcription was as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. qPCR was subsequently performed using the TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The temperature protocol for qPCR were as follows: 50°C for 2 min, 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec; and annealing/extension at 60°C for 60 sec. To analyze ZEB1 mRNA expression levels, total RNA was first reverse transcribed into cDNA using the PrimeScript RT Reagent kit (Takara Biotechnology, Co., Ltd., Dalian, China). The temperature protocol for reverse transcription was as follows: 37°C for 15 min and 85°C for 5 sec. qPCR was subsequently performed using the SYBR Premix Ex Taq™ kit (Takara Biotechnology, Co., Ltd.). The temperature protocol for qPCR was as follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. ZEB1 and miR-508 expression was quantified using the 2−ΔΔCq method and normalized to the internal reference gene GAPDH or U6 small nuclear RNA, respectively (26). The primer sequences used were as follows: miR-508 forward, 5′-TTCAAGAGACATGAGTGAC-3′ and reverse, 5′-TCTCTTGAACATGAGTGACG-3′; U6 forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; ZEB1 forward, 5′-AAGTGGCGGTAGATGGTA-3′ and reverse, 5′-TTGTAGCGACTGGATTTT-3′; and GAPDH forward, 5′-AACGGATTTGGTCGTATTG-3′ and reverse, 5′-GGAAGATGGTGATGGGATT-3′.
Cell proliferation assay
The proliferative ability of ccRCC cells was analyzed using the cell counting kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, transfected cells were collected and seeded into 96-well plates at a density of 3×103 cells/well and cultured for 0, 24, 48 and 72 h at 37°C. Following incubation, 10 µl CCK-8 reagent was then added to each well and the cells were incubated for an additional 2 h at 37°C. Cell proliferation was determined by measuring the absorbance at a wavelength of 450 nm using the SpectraMax Microplate® Spectrophotometer (Molecular Devices LLC, Sunnyvale, CA, USA).
Transwell invasion assay
ccRCC cell invasion was examined using an in vitro Transwell invasion chamber assay using Transwell™ polycarbonate membrane inserts for 24-well plates containing 8-µm pores (Corning Inc., Corning, NY, USA) precoated with Matrigel® (BD Biosciences, San Jose, CA, USA). Following transfection, 5×104 cells in serum-free DMEM were added to the upper chamber of the Transwell insert. In the lower chamber, 600 µl DMEM supplemented with 10% FBS was used as a chemo-attractant, and the plates were incubated at 37°C in a 5% CO2-humidified incubator for 24 h. Non-invasive cells remaining on the polycarbonate membrane were carefully removed and invaded cells were fixed with 100% methanol at room temperature for 30 min and stained with 0.1% crystal violet at room temperature for 30 min. The invasive capacity of ccRCC cells was quantified by counting the number of invaded cells in five randomly selected visual fields/chamber under an inverted light microscope (×200 magnification).
Bioinformatics analysis and dual-luciferase reporter assay
TargetScan (http://www.targetscan.org) and miRDB (http://mirdb.org) databases were used to identify putative targets of miR-508. ZEB1 was predicted as a potential target gene of miR-508. The wild-type (wt) and mutant (mut) fragments of the ZEB1 3′-UTR (synthesized by Shanghai GenePharma Co., Ltd.) were amplified by PCR and cloned into the firefly luciferase-expressing pmirGLO luciferase reporter plasmid (Promega Corporation, Madison, WI, USA). Cells were seeded into 24-well plates and incubated at 37°C in a CO2-humidified incubator for 24 h prior to transfection. Cells were subsequently co-transfected with miR-508 mimics or miR-NC and wt or mut 3′-UTR ZEB1 luciferase reporter plasmids using Lipofectamine® 2000. Following incubation for 48 h, luciferase activities were detected using the Dual-Luciferase Reporter assay system (Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activity was normalized to that of Renilla luciferase activity.
Western blot analysis
Total protein was extracted from tissues or cells using ice-cold radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with proteinase inhibitor cocktail (Roche Applied Science, Penzberg, Germany). Total protein was quantified using a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.) and equal quantities of protein (30 µg/lane) were separated via SDS-PAGE on a 10% gel. The separated proteins were transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) and blocked for 1 h at room temperature with 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20. The membranes were incubated with the following primary antibodies: Mouse anti-human monoclonal ZEB1 (dilution, 1:1,000; cat. no. sc-81428) or mouse anti-human monoclonal GAPDH (dilution, 1:1,000; cat. no. sc-47724; both Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Following primary incubation, the membranes were subsequently incubated with goat-anti mouse horseradish peroxidase-labeled secondary antibody (dilution, 1:5,000; cat. no. sc-516102; Santa Cruz Biotechnology, Inc.) at room temperature for 2 h. Protein bands were visualized using enhanced chemiluminescence detection reagent (EMD Millipore), according to the manufacturer's protocol. Protein expression was quantified by normalizing target protein expression to the GAPDH loading control. Quantity One software version 4.62 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for densitometry analysis.
Statistical analysis
The results are presented as the mean ± standard deviation from at least three independent experiments. All statistical analyses were performed using SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). The statistical significance of differences between two groups was analyzed using a two-tailed, paired Student's t-test. One-way analysis of variance followed by the Student-Newman-Keuls post hoc test was used to analyze differences among multiple groups. The association between miR-508 and ZEB1 mRNA expression in ccRCC tissue samples was determined using Spearman's correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-508 expression is downregulated in ccRCC tissue samples and cell lines
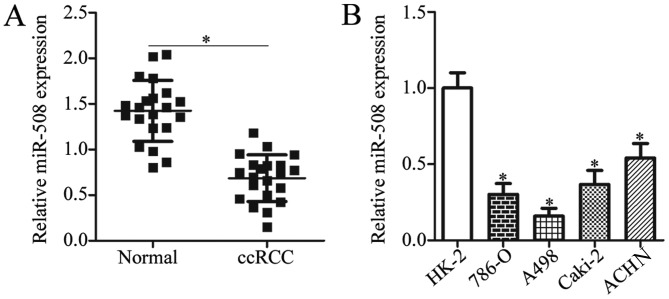
To investigate the expression pattern of miR-508 in ccRCC, the expression levels of miR-508 in ccRCC tissue samples and adjacent normal tissue samples from patients was determined by RT-qPCR analysis. The miR-508 expression level was significantly decreased in ccRCC tissue samples when compared with adjacent normal tissue samples (P<0.05; Fig. 1A). RT-qPCR was then used to examine miR-508 expression in two ccRCC cell lines (786-O and A498), two papillary RCC cell lines (Caki-2 and ACHN), and the normal human renal cell line HK-2. The expression of miR-508 was significantly decreased in ccRCC and papillary RCC cell lines when compared with the normal human renal cell line (P<0.05; Fig. 1B). These results suggest that downregulation of miR-508 may be associated with ccRCC progression.
Figure 1.
Downregulation of miR-508 in ccRCC tissue samples and cell lines. (A) The relative expression level of miR-508 was determined by RT-qPCR in ccRCC and adjacent normal tissue samples from patients with ccRCC. *P<0.05 vs. Normal. (B) The relative expression level of miR-508 was determined by RT-qPCR in 786-O, and A498 ccRCC cell lines, the Caki-2 and ACHN papillary RCC cell line and the normal human renal cell line, HK-2. *P<0.05 vs. HK-2. miR, microRNA; ccRCC, clear cell renal cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-508 overexpression suppresses the proliferation and invasion of ccRCC cells
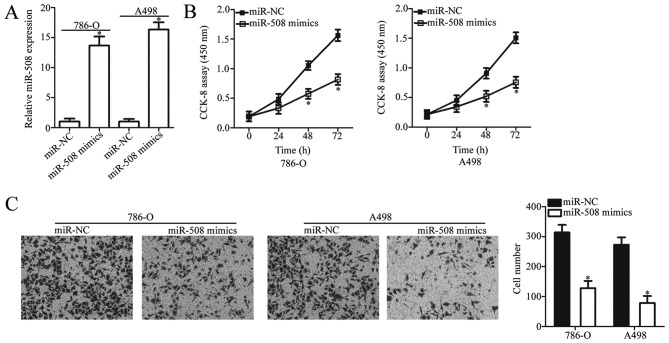
To investigate the biological function of miR-508 in ccRCC progression, ccRCC cell lines 786-O and A498 (as they exhibited the lowest miR-508 expression) were transfected with miR-508 mimics or miR-NC. The relative expression of miR-508 was significantly increased in ccRCC cells following transfection with miR-508 mimics compared with miR-NC (P<0.05; Fig. 2A). A CCK-8 assay was then used to examine the effect of miR-508 overexpression on ccRCC cell proliferation. The results demonstrated that miR-508 overexpression significantly inhibited the proliferative ability of 786-O and A498 cells when compared with the negative controls (P<0.05; Fig. 2B). Subsequently, the in vitro Transwell invasion assay demonstrated that miR-508 overexpression significantly inhibited the invasive capacity of 786-O and A498 cells compared with the negative controls (P<0.05; Fig. 2C). Taken together, these results suggest that miR-508 may serve a tumor-suppressive role in the development of ccRCC.
Figure 2.
miR-508 overexpression suppresses the proliferation and invasion of 786-O and A498 cells. (A) The relative expression level of miR-508 in 786-O and A498 cells was determined by reverse transcription-quantitative polymerase chain reaction analysis at 48 h following transfection with miR-508 mimics or miR-NC. Cell (B) proliferation and (C) invasion were examined using CCK-8 and in vitro Transwell invasion assays, respectively, in 786-O and A498 cells at 48 h following transfection with miR-508 mimics or miR-NC. *P<0.05 vs. miR-NC. miR, microRNA; CCK-8, cell counting kit-8; NC, negative control.
ZEB1 is a direct target gene of miR-508 in ccRCC cells
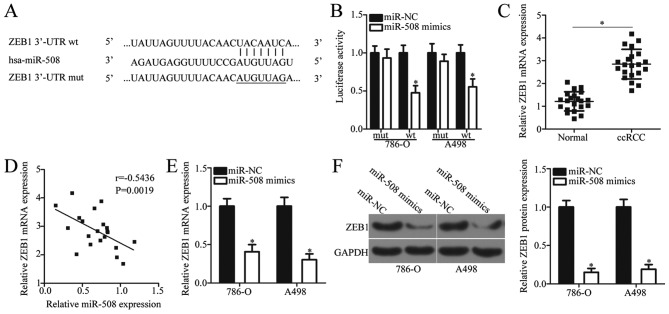
It is well established that miRNAs directly target mRNAs of target genes, thereby exerting their role as post-transcriptional regulators (12). To further investigate the mechanisms underlying the tumor suppressive role of miR-508 in ccRCC in the present study, putative targets of miR-508 were examined by bioinformatics analysis. TargetScan and miRDB databases identified ZEB1, which contains a 3′-UTR sequence complementary to the seed sequence of miR-508 (Fig. 3A). Therefore, ZEB1 was selected for verification using a dual-luciferase reporter assay. This assay was used to confirm whether miR-508 binds directly to the partially complimentary sequence within the 3′-UTR of ZEB1 in ccRCC cells. The results demonstrated that miR-508 overexpression significantly decreased the luciferase activity of the reporter plasmid containing the wt miR-508 binding site, whereas co-transfection with the mut miR-508 binding site plasmid demonstrated no effect on luciferase activity (P<0.05; Fig. 3B).
Figure 3.
ZEB1 is a direct target gene of miR-508 in ccRCC cells. (A) Bioinformatics analysis was used to predict the miR-508 binding site in the wt 3′-UTR of ZEB1. (B) Luciferase reporter assays were performed in 786-O and A498 cells following co-transfection with luciferase reporter plasmids containing the wt or mut 3′-UTR of ZEB1 and miR-508 mimics or miR-NC. *P<0.05 vs. miR-NC. (C) The relative mRNA expression level of ZEB1 was determined by RT-qPCR in ccRCC and adjacent normal tissue samples from patients with ccRCC. *P<0.05 vs. Normal. (D) An inverse correlation between miR-508 and ZEB1 mRNA expression levels in ccRCC tissue samples from patients with ccRCC was identified. (E) The relative mRNA expression level of ZEB1 was determined by RT-qPCR in 786-O and A498 cells at 48 h following transfection with miR-508 mimics or miR-NC. *P<0.05 vs. miR-NC. (F) The relative protein expression level of ZEB1 was determined by western blot analysis in 786-O and A498 cells following 48-h transfection with miR-508 mimics or miR-NC. *P<0.05 vs. miR-NC. ZEB1, zinc finger E-box binding homeobox 1; miR, microRNA; ccRCC, clear cell renal cell carcinoma; UTR, untranslated region; wt, wild-type; mut, mutant; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control.
To further understand the association between miR-508 and ZEB1 in ccRCC, the expression of ZEB1 was analyzed using RT-qPCR in ccRCC and normal adjacent tissue samples from patients with ccRCC. The mRNA levels of ZEB1 were significantly increased in ccRCC tissue samples when compared with adjacent normal tissue samples (P<0.05; Fig. 3C). In addition, Spearman's correlation analysis indicated a statistically significant negative correlation between miR-508 and ZEB1 mRNA expression levels in ccRCC tissue samples (r=−0.5436, P=0.0019; Fig. 3D). To investigate whether ZEB1 expression was regulated by miR-508 in ccRCC cells, ZEB1 expression levels were determined by RT-qPCR in 786-O and A498 cells transfected with miR-508 mimics or miR-NC. The mRNA and protein expression levels of ZEB1 significantly decreased following miR-508 overexpression compared with the negative controls (P<0.05; Fig. 3E and F). These results suggest that ZEB1 may be a direct target of miR-508 in ccRCC cells.
Inhibition of ZEB1 suppresses ccRCC cell proliferation and invasion
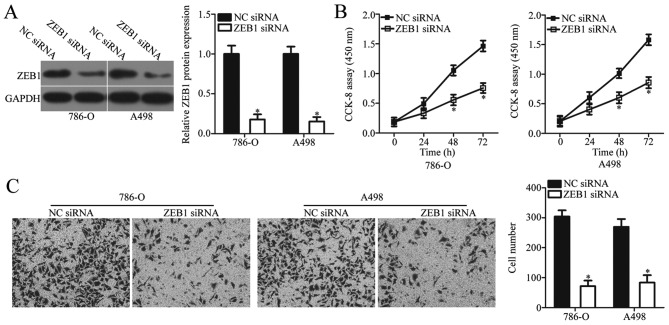
To examine the biological role of ZEB1 in ccRCC, 786-O and A498 cells were transfected with ZEB1 siRNA or NC siRNA. Endogenous ZEB1 protein expression was significantly decreased in ccRCC cells following transfection with ZEB1 siRNA when compared with NC siRNA (P<0.05; Fig. 4A). CCK-8 and in vitro Transwell invasion assays were used to examine the effect of ZEB1 knockdown on ccRCC cell proliferation and invasion, respectively. The results demonstrated that ZEB1 knockdown significantly inhibited the proliferative and invasive ability of 786-O and A498 cells when compared with the negative controls (P<0.05; Fig. 4B and C). Taken together, these results demonstrated that ZEB1 knockdown exerts a functional role similar to that of miR-508 overexpression in ccRCC cells, which suggests that ZEB1 may be a functional target of miR-508 in ccRCC.
Figure 4.
Inhibition of ZEB1 suppresses 786-O and A498 cell proliferation and invasion. (A) The relative protein expression level of ZEB1 was determined by western blot analysis in 786-O and A498 cells at 48 h following transfection with ZEB1 siRNA or NC siRNA. Cell (B) proliferation and (C) invasion were examined using CCK-8 and in vitro Transwell invasion assays, respectively, in 786-O and A498 cells at 48 h following transfection with ZEB1 siRNA or NC siRNA. *P<0.05 vs. NC siRNA. ZEB1, zinc finger E-box binding homeobox 1; siRNA, small interfering RNA; NC, negative control.
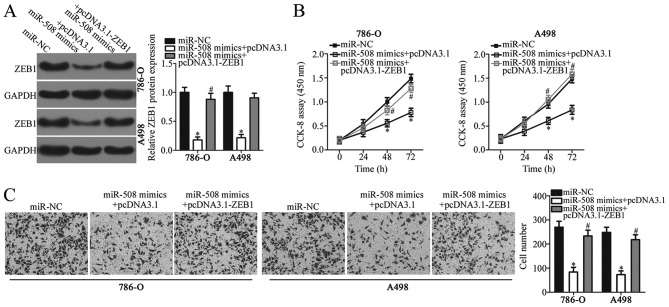
Restoration of ZEB1 expression reverses the suppressive effects of miR-508 on the malignant phenotype of ccRCC cells
As ZEB1 was identified as a direct target of miR-508 in ccRCC, rescue assays were performed to determine whether ZEB1 knockdown was responsible for the miR-508-induced inhibitory effects on ccRCC cell proliferation and invasion. ccRCC cell lines 786-O and A498 were co-transfected with miR-508 mimics and pcDNA3.1 or pcDNA3.1-ZEB1. Western blot analysis confirmed that the relative protein expression levels of ZEB1 were significantly increased in ccRCC cells following co-transfection with miR-508 mimics and pcDNA3.1-ZEB1 compared with miR-508 mimics and the pcDNA3.1 negative control (P<0.05; Fig. 5A). Furthermore, functional analysis demonstrated that restoration of ZEB1 expression significantly reversed the inhibitory effects of miR-508 on ccRCC cell proliferation and invasion (P<0.05; Fig. 5B and C). These results suggest that miR-508 may serve a tumor suppressive role in ccRCC and that the inhibitory effects of this miRNA may be, in part, mediated by regulating ZEB1 expression.
Figure 5.
Restoration of ZEB1 expression reverses the suppressive effects of miR-508 on 786-O and A498 cell proliferation and invasion. (A) The relative protein expression level of ZEB1 was determined by western blot analysis in 786-O and A498 cells following co-transfection with miR-508 and pcDNA3.1 or pcDNA3.1-ZEB1 lacking the 3′-UTR of ZEB1. Cell (B) proliferation and (C) invasion were examined using CCK-8 and in vitro Transwell invasion assays, respectively, in 786-O and A498 cells following co-transfection with miR-508 and pcDNA3.1 or pcDNA3.1-ZEB1 lacking the 3′-UTR of ZEB1. *P<0.05 vs. miR-NC; #P<0.05 vs. miR-508 mimics + pcDNA3.1. ZEB1, zinc finger E-box binding homeobox 1; miR, microRNA; UTR, untranslated region; NC, negative control.
Discussion
A number of recent studies have identified a variety of deregulated miRNAs in ccRCC, including miR-224 (27), miR-502 (28), miR-543 (18) and miR-645 (29). miRNAs are involved in several biological and pathological processes, including ccRCC occurrence and development (30–32). Previous studies have also identified several deregulated miRNAs in ccRCC, whereby oncogenic miRNAs are upregulated and tumor suppressor miRNAs are downregulated (17,33). Therefore, understanding the expression pattern and biological function of miRNAs in ccRCC, as well as the underlying mechanisms of miRNAs in the development and progression of ccRCC, may be useful for the identification of novel therapeutic targets in the treatment of patients with ccRCC. In the current study, miR-508 expression in ccRCC tissue samples and cell lines was examined and the regulatory effects of miR-508 in the development of ccRCC in vitro were examined. Furthermore, the underlying molecular mechanisms of miR-508 in ccRCC cells were investigated. The current study identified miR-508 as a putative novel biomarker in the diagnosis of ccRCC, as well as a potentially novel and efficient therapeutic target for the treatment of patients with ccRCC.
Previous studies have demonstrated that miR-508 expression is downregulated in ovarian cancer (22) and colorectal cancer (23) compared with normal tissues of the same type. By contrast, miR-508 was observed to be upregulated in esophageal squamous cell carcinoma compared with normal esophageal tissues (24). Patients with esophageal squamous cell carcinoma expressing high levels of miR-508 exhibited worse survival rates when compared with patients with low miR-508 expression (24). However, the expression status of miR-508 in ccRCC requires further investigation. In the present study, miR-508 expression was significantly decreased in ccRCC tissue samples and cell lines when compared with adjacent normal tissue samples and the normal human renal cell line HK-2, respectively. These conflicting results suggest that miR-508 may exert tissue-specific expression patterns, and miR-508 may therefore present a potential diagnostic biomarker for patients with these types of cancer.
Previous studies have demonstrated that aberrant miR-508 expression may contribute to the development and progression of several types of human cancer (22–24). Upregulation of miR-508 suppressed cell proliferation, metastasis and epithelial-to-mesenchymal transition in ovarian cancer via the mitogen-activated protein kinase 1 signaling pathway (22). In addition, Yan et al (23) reported that ectopic expression of miR-508 in colorectal cancer cells attenuated epithelial-to-mesenchymal transition, stemness and metastasis in vitro and in vivo. By contrast, miR-508 exerts an oncogenic role in esophageal squamous cell carcinoma by promoting tumorigenicity both in vivo and in vitro (24). These studies suggest that the role of miR-508 in cancer development and progression may be tissue specific. miR-508 may function as tumour suppressor or promoter in different types of human cancer, depending on the characteristics of corresponding target genes (34). However, the potential role of miR-508 in ccRCC remains unknown. In the current study, functional experiments indicated that miR-508 overexpression inhibited the proliferation and invasion of ccRCC cells. These findings suggest that miR-508 may serve as a potential therapeutic target in the treatment of patients with ccRCC.
miRNAs regulate several biological and pathological processes, including tumorigenesis, by binding to specific sites within the 3′-UTR of target mRNA sequences to inhibit their translation and expression (12). In the current study, the underlying mechanism of miR-508 in ccRCC cells was investigated. Bioinformatics analysis identified ZEB1, which contains a 3′-UTR sequence complementary to the seed sequence of miR-508. Therefore, ZEB1 was considered a potential target gene of miR-508 in the present study. A dual-luciferase reporter assay was used to confirm a direct interaction between the 3′-UTR of ZEB1 and miR-508 in ccRCC cells. RT-qPCR and western blot analyses revealed that the mRNA and protein expression levels of ZEB1 were negatively regulated by miR-508 in ccRCC cells. In addition, the current study demonstrated that ZEB1 expression was significantly increased in ccRCC tissue samples, and ZEB1 expression was inversely correlated with miR-508 expression in these tissue samples. Finally, restoration of ZEB1 expression reversed the inhibitory effect of miR-508 on the malignant phenotype of ccRCC cells. Taken together, these results suggest that ZEB1 is a direct and functional downstream target of miR-508 in ccRCC cells.
ZEB1, located on the short arm of human chromosome 10, is a member of the ZEB family of transcription factors (35). Previous studies have demonstrated that ZEB1 is upregulated in several types of cancer, including lung (36), gastric (37), colorectal (38) and endometrial cancers (39). Overexpression of ZEB1 has also been identified in ccRCC, and ZEB1 overexpression was significantly correlated with tumor grade, tumor, node and metastasis stage, lymph node metastasis and distant metastases (40). In addition, patients with ccRCC expressing high ZEB1 expression levels exhibited worse overall and progression-free survival when compared with low ZEB1 expression (40). Furthermore, ZEB1 was demonstrated to be involved in several biological processes in ccRCC development and progression (41). The current study demonstrated that miR-508 directly targets ZEB1 and suppresses ccRCC cell proliferation and invasion. The miR-508-ZEB1 axis may therefore be a novel and efficient therapeutic target, which could be used to inhibit the rapid growth and metastasis of ccRCC.
In conclusion, to the best of the authors' knowledge, the current study is the first to demonstrate that miR-508 is downregulated in ccRCC and that miR-508 may inhibit the development of ccRCC by directly targeting ZEB1. The miR-508-ZEB1 axis may present a potential therapeutic target in the treatment of patients with ccRCC patients. However, the current study has several limitations that will need to be addressed in future studies. The effect of miR-508 in cell apoptosis, in vivo tumor growth, angiogenesis, energy metabolism and drug resistance were not examined. Due to the relatively small sample size, the correlation between clinicopathological features and miR-508 expression in ccRCC tissue samples was also not examined. Furthermore, future studies will be required to validate miR-508 as a potential biomarker in the early diagnosis and prognosis of patients with ccRCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundations of China (grant no. 31700720), Project of Chengguan District Science and Technology Bureau of Lanzhou city (grant no. 2017SHFZ0029) and the Doctoral Research Fund of the Second Hospital of Lanzhou University (grant no. ynbskyjj2015-1-13).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZY and WW designed the study. WW and WH performed RT-qPCR, CCK-8 and in vitro Transwell invasion assays. YW performed the dual-luciferase reporter assay. JY performed western blotting and statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Lanzhou University Second Hospital (Lanzhou, China). The present study was performed in accordance with the Declaration of Helsinki and the guidelines of the Ethics Committee of Lanzhou University Second Hospital. Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu Y, Han X, Yu Y, Ding Y, Ni C, Liu W, Hou X, Li Z, Hou J, Shen D, et al. A genetic polymorphism affects the risk and prognosis of renal cell carcinoma: Association with follistatin-like protein 1 expression. Sci Rep. 2016;6:26689. doi: 10.1038/srep26689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia JA, Cowey CL, Godley PA. Renal cell carcinoma. Curr Opin Oncol. 2009;21:266–271. doi: 10.1097/CCO.0b013e32832a05c8. [DOI] [PubMed] [Google Scholar]
- 3.Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, De La Taille A, Tostain J, Artibani W, Abbou CC, et al. Prognostic value of histologic subtypes in renal cell carcinoma: A multicenter experience. J Clin Oncol. 2005;23:2763–2771. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 4.Nerich V, Hugues M, Paillard MJ, Borowski L, Nai T, Stein U, Nguyen Tan Hon T, Montcuquet P, Maurina T, Mouillet G, et al. Clinical impact of targeted therapies in patients with metastatic clear-cell renal cell carcinoma. Onco Targets Ther. 2014;7:365–374. doi: 10.2147/OTT.S56370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu D, Li M, Wang L, Zhou Y, Zhou J, Pan H, Qu P. microRNA145 inhibits cell proliferation, migration and invasion by targeting matrix metallopeptidase-11 in renal cell carcinoma. Mol Med Rep. 2014;10:393–398. doi: 10.3892/mmr.2014.2149. [DOI] [PubMed] [Google Scholar]
- 6.Crispen PL, Breau RH, Allmer C, Lohse CM, Cheville JC, Leibovich BC, Blute ML. Lymph node dissection at the time of radical nephrectomy for high-risk clear cell renal cell carcinoma: Indications and recommendations for surgical templates. Eur Urol. 2011;59:18–23. doi: 10.1016/j.eururo.2010.08.042. [DOI] [PubMed] [Google Scholar]
- 7.Flanigan RC, Campbell SC, Clark JI, Picken MM. Metastatic renal cell carcinoma. Curr Treat Options Oncol. 2003;4:385–390. doi: 10.1007/s11864-003-0039-2. [DOI] [PubMed] [Google Scholar]
- 8.Breda A, Konijeti R, Lam JS. Patterns of recurrence and surveillance strategies for renal cell carcinoma following surgical resection. Expert Rev Anticancer Ther. 2007;7:847–862. doi: 10.1586/14737140.7.6.847. [DOI] [PubMed] [Google Scholar]
- 9.Yang YQ, Chen J. Predictive role of vascular endothelial growth factor polymorphisms in the survival of renal cell carcinoma patients. Genet Mol Res. 2014;13:5011–5017. doi: 10.4238/2014.July.4.16. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 13.Bryzgunova OE, Konoshenko MY, Laktionov PP. MicroRNA-guided gene expression in prostate cancer: Literature and database overview. J Gene Med. 2018;20:e3016. doi: 10.1002/jgm.3016. [DOI] [PubMed] [Google Scholar]
- 14.Pratap P, Raza ST, Abbas S, Mahdi F. MicroRNA-associated carcinogenesis in lung carcinoma. J Cancer Res Ther. 2018;14:249–254. doi: 10.4103/0973-1482.187283. [DOI] [PubMed] [Google Scholar]
- 15.Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133. doi: 10.1016/j.gde.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Lou L, Zhong K, Wan L. MicroRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. Exp Biol Med (Maywood) 2017;242:1299–1305. doi: 10.1177/1535370217701625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al. Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem. 2017;43:2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Ma J, Tang Q, Zhang W, Fu Q, Sun J, Wang H, Song B. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Krüppel-like factor 6. Biomed Pharmacother. 2018;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 19.He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233:4458–4465. doi: 10.1002/jcp.26347. [DOI] [PubMed] [Google Scholar]
- 20.Jaswani P, Prakash S, Dhar A, Sharma RK, Prasad N, Agrawal S. MicroRNAs involvement in renal pathophysiology: A bird's eye view. Indian J Nephrol. 2017;27:337–341. doi: 10.4103/ijn.IJN_264_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellinger J, Gevensleben H, Muller SC, Dietrich D. The emerging role of non-coding circulating RNA as a biomarker in renal cell carcinoma. Expert Rev Mol Diagn. 2016;16:1059–1065. doi: 10.1080/14737159.2016.1239531. [DOI] [PubMed] [Google Scholar]
- 22.Hong L, Wang Y, Chen W, Yang S. MicroRNA-508 suppresses epithelial-mesenchymal transition, migration, and invasion of ovarian cancer cells through the MAPK1/ERK signaling pathway. J Cell Biochem. 2018;119:7431–7440. doi: 10.1002/jcb.27052. [DOI] [PubMed] [Google Scholar]
- 23.Yan TT, Ren LL, Shen CQ, Wang ZH, Yu YN, Liang Q, Tang JY, Chen YX, Sun DF, Zgodzinski W, et al. miR-508 defines the stem-like/mesenchymal subtype in colorectal cancer. Cancer Res. 2018;78:1751–1765. doi: 10.1158/0008-5472.CAN-17-2101. [DOI] [PubMed] [Google Scholar]
- 24.Lin C, Liu A, Zhu J, Zhang X, Wu G, Ren P, Wu J, Li M, Li J, Song L. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat Commun. 2014;5:4620. doi: 10.1038/ncomms5620. [DOI] [PubMed] [Google Scholar]
- 25.Zhai Q, Zhou L, Zhao C, Wan J, Yu Z, Guo X, Qin J, Chen J, Lu R. Identification of miR-508-3p and miR-509-3p that are associated with cell invasion and migration and involved in the apoptosis of renal cell carcinoma. Biochem Biophys Res Commun. 2012;419:621–626. doi: 10.1016/j.bbrc.2012.02.060. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Fujii N, Hirata H, Ueno K, Mori J, Oka S, Shimizu K, Kawai Y, Inoue R, Yamamoto Y, Matsumoto H, et al. Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget. 2017;8:109877–109888. doi: 10.18632/oncotarget.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Guo Z, Xu J, Wang J, Zhang J, Cui L, Zhang H, Liu Y, Bai Y. miR-502-mediated histone methyltransferase SET8 expression is associated with clear cell renal cell carcinoma risk. Oncol Lett. 2017;14:7131–7138. doi: 10.3892/ol.2017.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Shu Y, Yu Q, Shen W. MicroRNA-645 promotes cell metastasis and proliferation of renal clear cell carcinoma by targeting GK5. Eur Rev Med Pharmacol Sci. 2017;21:4557–4565. [PubMed] [Google Scholar]
- 30.Ding D, Zhang Y, Wen L, Fu J, Bai X, Fan Y, Lin Y, Dai H, Li Q, Zhang Y, An R. MiR-367 regulates cell proliferation and metastasis by targeting metastasis-associated protein 3 (MTA3) in clear-cell renal cell carcinoma. Oncotarget. 2017;8:63084–63095. doi: 10.18632/oncotarget.18647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang Y, Bao X, Du Q, Luo G, Liu K, et al. miR-122 promotes metastasis of clear-cell renal cell carcinoma by downregulating Dicer. Int J Cancer. 2018;142:547–560. doi: 10.1002/ijc.31050. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Wu L, Wang A, Xu Y, Luo X, Liu X, Hua Y, Zhang D, Wu S, Lin T, et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res. 2017;9:3611–3622. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Cai L, Liu J, Wang G, Li H, Wang X, Xu W, Ren M, Feng L, Liu P, Zhang C. MicroRNA-30a-5p inhibits the growth of renal cell carcinoma by modulating GRP78 expression. Cell Physiol Biochem. 2017;43:2405–2419. doi: 10.1159/000484394. [DOI] [PubMed] [Google Scholar]
- 34.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol. 2012;105:830–834. doi: 10.1002/jso.23012. [DOI] [PubMed] [Google Scholar]
- 36.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–3235. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia B, Liu H, Kong Q, Li B. Overexpression of ZEB1 associated with metastasis and invasion in patients with gastric carcinoma. Mol Cell Biochem. 2012;366:223–229. doi: 10.1007/s11010-012-1299-6. [DOI] [PubMed] [Google Scholar]
- 38.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M, Spoelstra NS, Jean A, Howe E, Torkko KC, Clark HR, Darling DS, Shroyer KR, Horwitz KB, Broaddus RR, Richer JK. ZEB1 expression in type I vs. type II endometrial cancers: A marker of aggressive disease. Mod Pathol. 2008;21:912–923. doi: 10.1038/modpathol.2008.82. [DOI] [PubMed] [Google Scholar]
- 40.Harb OA, Elfeky MA, El Shafaay BS, Taha HF, Osman G, Harera IS, Gertallah LM, Abdelmonem DM, Embaby A. SPOP, ZEB-1 and E-cadherin expression in clear cell renal cell carcinoma (cc-RCC): Clinicopathological and prognostic significance. Pathophysiology. 2018;25:335–345. doi: 10.1016/j.pathophys.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Hong Z, Chai Y, Liu Z, Du Y, Li Q, Liu Q. CSN5 promotes renal cell carcinoma metastasis and EMT by inhibiting ZEB1 degradation. Biochem Biophys Res Commun. 2017;488:101–108. doi: 10.1016/j.bbrc.2017.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.