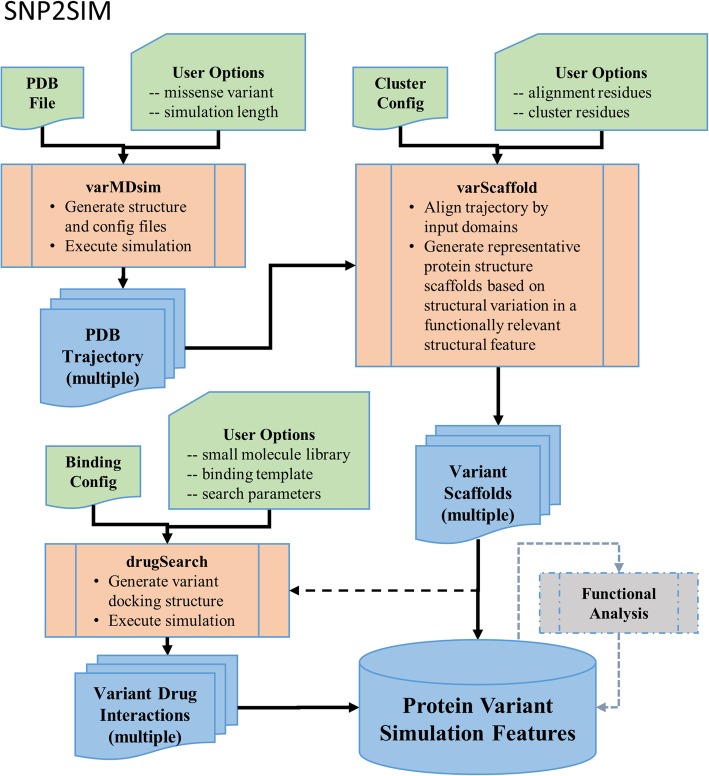
Fig. 1.
The SNP2SIM workflow contains 3 functional modules (shown in orange) that execute all atom molecular dynamics of protein structure variants using NAMD and VMD (varMDsim), clusters the resulting trajectories into a set of structures that represent the conformational dynamics of the binding interface (varScaffold), and predicts the binding interactions of low molecular weight ligands using AutoDock Vina (drugSearch). The input for each module (green) control their configuration, providing a way to standardize simulation parameters across parallel computational infrastructures. The resulting structural datasets (blue) can be used to analyze protein:ligand interactions and enables large scale investigations into the functional consequences of protein sequence variation