Fig. 2.
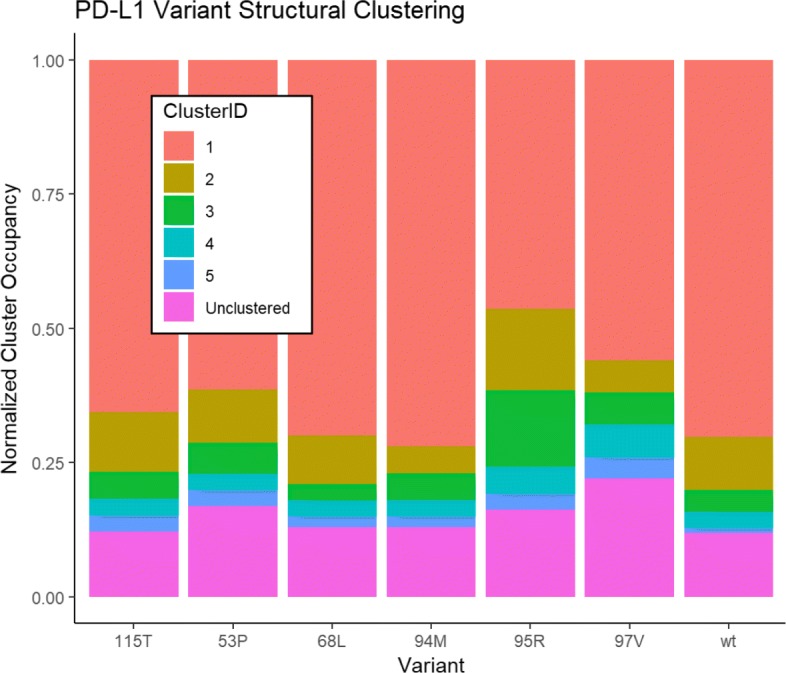
The breakdown of the results from the varScaffold module of the SNP2SIM workflow show the variation induced changes to the organization of the PD-L1 binding residues in the simulated structures. The clusters are ranked by the total number of MD conformations that fall within the user supplied RMSD threshold, and the remaining structures that are not assigned to the top 5 clusters are given the “Unclustered” designation. A representative structure from each cluster that contains at least 10% of the total structures derived from the simulated trajectories are used to create a representative scaffold for drug binding
