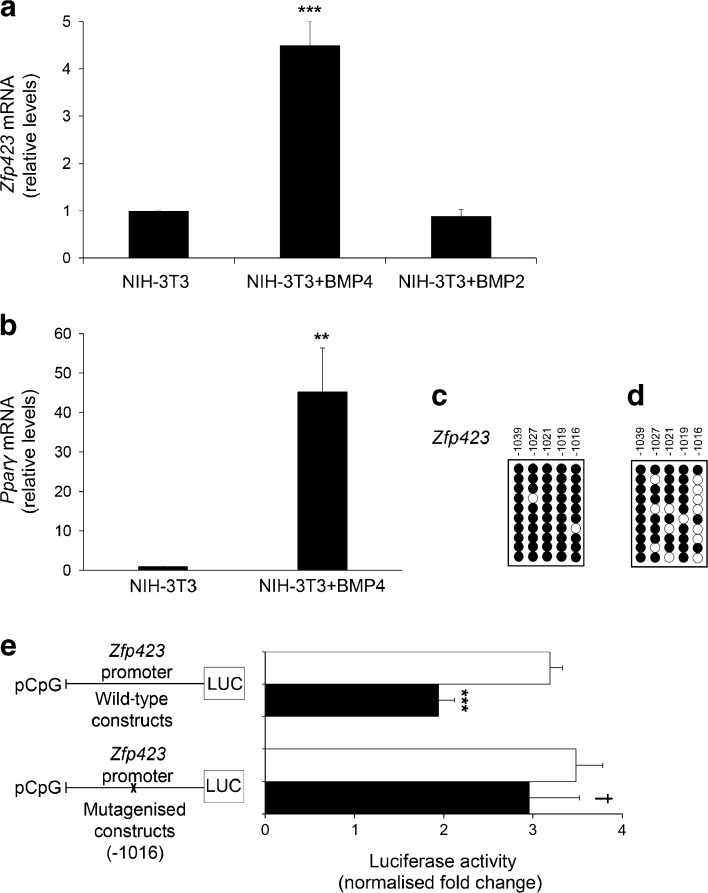
Fig. 5.
Effect of BMP4 on Zfp423 mRNA expression and promoter methylation in NIH-3T3 cells. Cells were cultured in medium supplemented with 50 ng/ml BMP4 and 100 ng/ml BMP2. (a, b) Expression of Zfp423 and Pparγ mRNA was measured by qPCR. Results are the mean ± SD from three independent experiments. Statistical significance was analysed by two-tailed Student’s t tests (**p < 0.01, ***p < 0.001 vs NIH-3T3). (c, d) Bisulphite sequencing analysis of the DNA methylation status and percentage of the methylated −1016 CpG position of the Zfp423 promoter in NIH-3T3 cells on exposure to BMP4 (methylation −1016 CpG: 90 ± 10% NIH-3T3 [c], 30 ± 10% NIH-3T3 + BMP4 [d]). Each PCR product was subcloned, and ten clones were analysed by bisulphite sequencing. The methylation profile of each CpG site of the Zfp423 promoter, either methylated (black circles) or unmethylated (white circles), was aligned to their sequence position. Results are the mean ± SD from three independent experiments. Statistical significance was tested by two-tailed Student’s t tests (p < 0.01 vs NIH-3T3 without BMP4). (e) Effect of mutagenesis at the −1016 CpG position of the Zfp423 promoter. Disruption of CpG was performed by site-directed mutagenesis as described in Methods. Wild-type and mutated plasmids were treated with the DNA-methylase M.SssI and transfected into NIH-3T3 cells (black bars). White bars, untreated plasmids. Luciferase activity was normalised to Renilla luciferase activity. Error bars represent SD from three replicates. Statistical significance was tested by two-tailed Student’s t tests (***p < 0.001 vs wild-type unmethylated; † p < 0.05 vs wild-type methylated). LUC, luciferase reporter