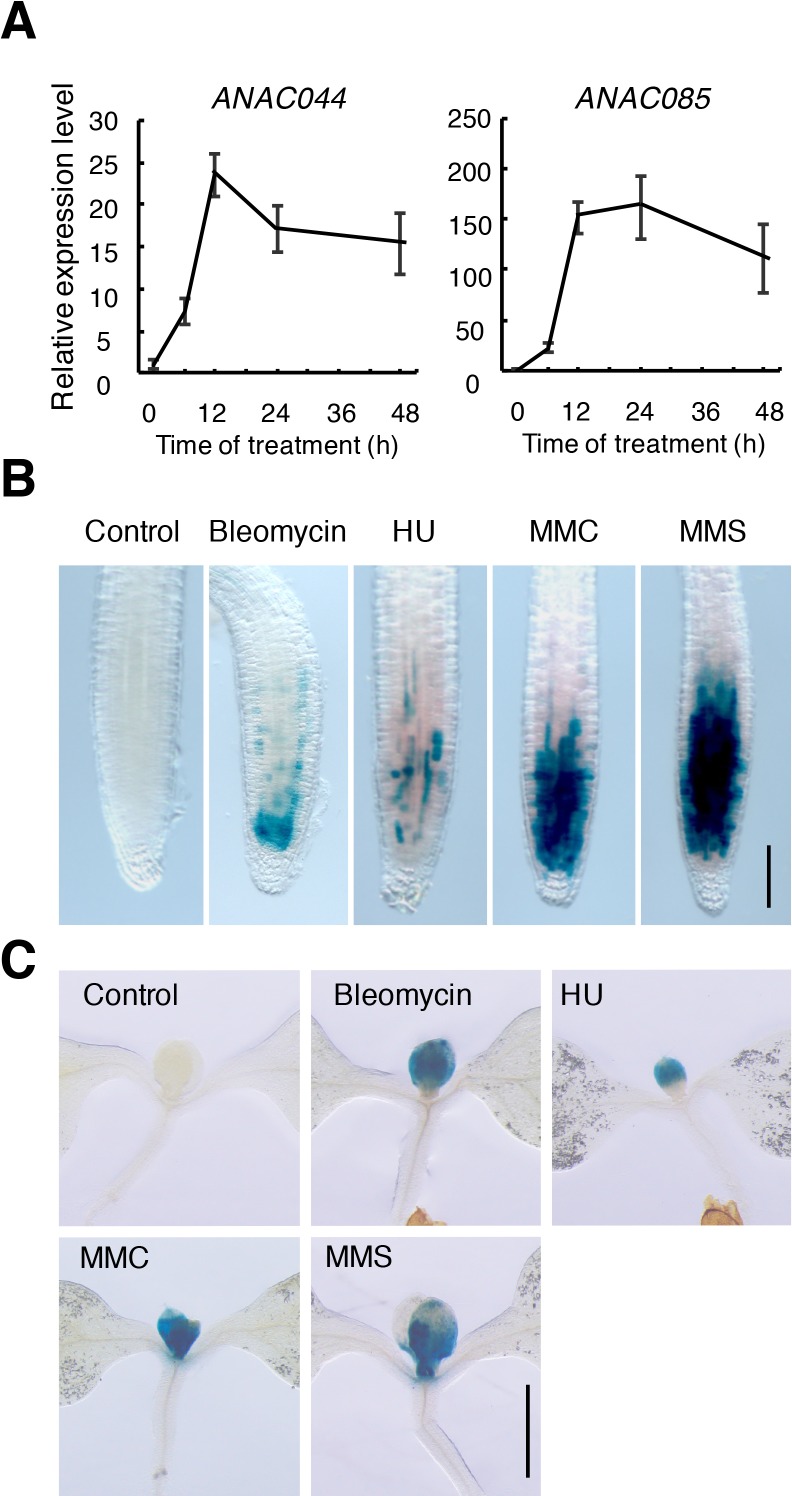
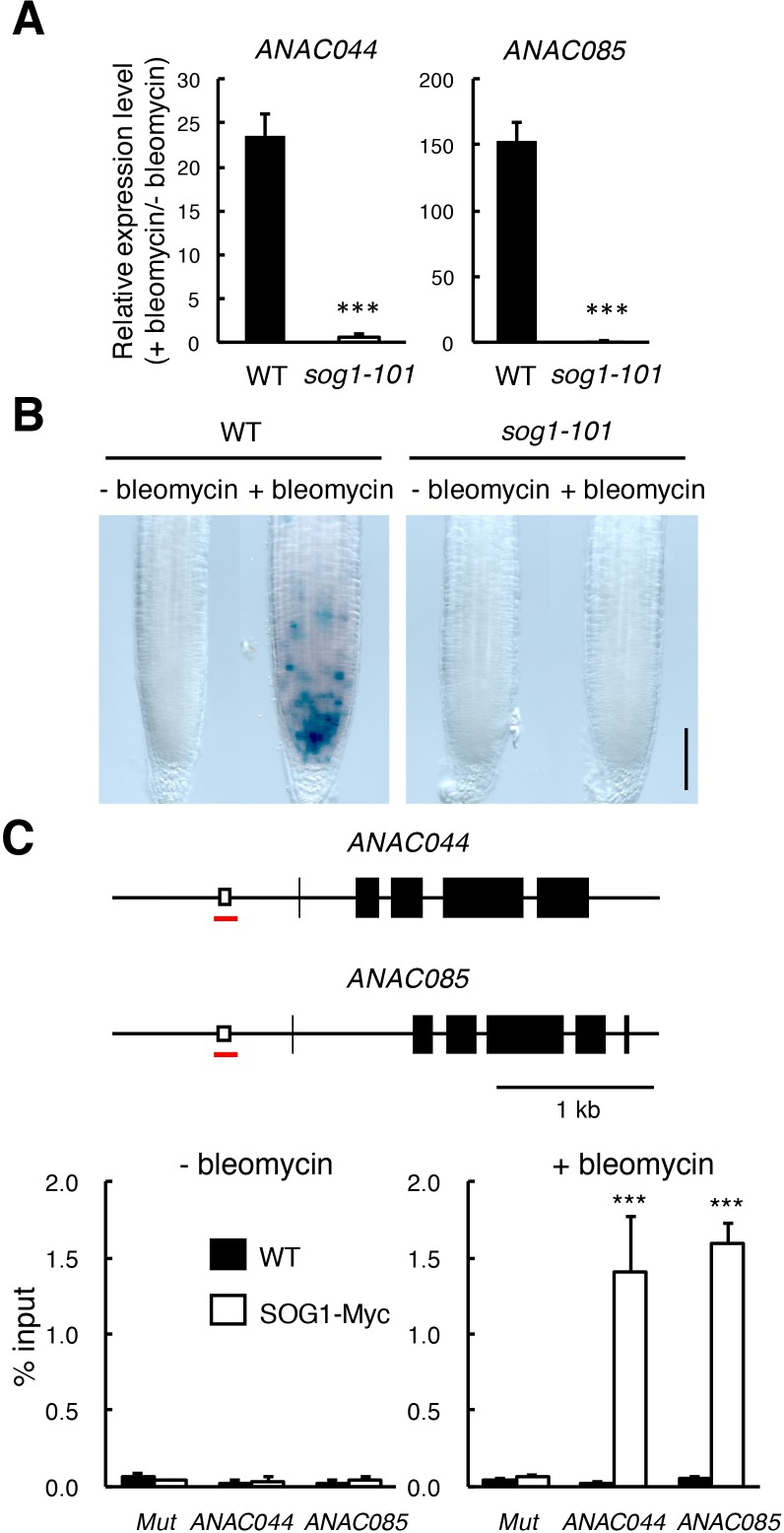
(A) Transcript levels of ANAC044 and ANAC085 after bleomycin treatment. Five-day-old WT seedlings were treated with 0.6 µg/ml bleomycin for 0, 6, 12, 24 or 48 hr. The mRNA levels were normalized to that of ACTIN2 and are indicated as relative values, with that for 0 hr set to 1. Data are presented as mean ±SD (n = 3). (B, C) Five-day-old seedlings harbouring ProANAC044:GUS were transferred to medium supplemented with or without 0.6 µg/ml bleomycin, 1.5 mM hydroxyurea (HU), 3.3 µg/ml mitomycin C (MMC) or 80 ppm methyl methanesulfonate (MMS), and grown for 24 hr. GUS-stained seedlings were observed for the root tip (B) and shoot apex (C). Bars = 100 µm (B) and 1 mm (C).
Figure 1—figure supplement 1—source data 1. Source data.