Abstract
TP53 is the most frequently mutated or deleted gene in triple negative breast cancer (TNBC). Both the loss of TP53 and the lack of targeted therapy are significantly correlated with poor clinical outcomes, making TNBC the only type of breast cancer that has no approved targeted therapies. Through in silico analysis, we identified POLR2A in the TP53-neighboring region as a collateral vulnerability target in TNBC tumours, suggesting that its inhibition via small interfering RNA (siRNA) may be an amenable approach for TNBC targeted treatment. To enhance bioavailability and improve endo/lysosomal escape of siRNA, we designed pH-activated nanoparticles for augmented cytosolic delivery of POLR2A siRNA (siPol2). Suppression of POLR2A expression with the siPol2-laden nanoparticles (siPol2@NPs) leads to enhanced growth reduction of tumours characterised by hemizygous POLR2A loss. These results demonstrate the potential of the pH-responsive nanoparticle and the precise POLR2A targeted therapy in TNBC harbouring the common TP53 genomic alteration.
Triple negative breast cancer (TNBC) is negative for the expression of oestrogen and progesterone receptors, and absent of human epidermal growth factor receptor 2 (HER2) overexpression1–3. These receptors are molecular targets for treating breast cancer1–3. As a result, other than olaparib, a poly(ADP-ribose) polymerase inhibitor that can benefit a small subset of TNBC patients with BRCA mutation, no approved targeted therapies are available for most TNBC patients. Standard chemotherapy is the only approved option, but it is ineffective with undesired side effects4,5. Therefore, new targeted therapies are critically needed for TNBC.
Cancer genomes are characterized by the accumulation of somatic genetic alterations within a cell, such as inactivation of tumour suppressor genes6–8. TP53 is the most frequently deleted or mutated tumour suppressor gene in TNBC3,9,10, which results in the loss of p53’s tumour suppressor function11,12. Although restoration of p53 activity is a promising strategy and tremendous efforts have been made to harness it as an anticancer approach, no such therapy has been translated into the clinic owing to the complexity of p53 signaling13. Because genomic alterations are large regional events, most cancers that exhibit copy number loss of tumour suppressor genes, especially TP53, also show loss of essential neighbouring genes14. POLR2A is an essential neighbouring gene of TP53 that encodes the largest subunit of RNA polymerase II complex15. Although hemizygous (partial) loss of POLR2A (POLR2Aloss) has minimal impact on cells because one allele of POLR2A is sufficient to maintain cell survival, cancer cells containing such genomic defect should be more vulnerable than normal cells to the inhibition of POLR2A. Therefore, we propose to precisely target POLR2A instead of TP53 for treating TNBC harbouring hemizygous loss of TP53 (TP53loss).
RNA interference (RNAi) with small interfering RNA (siRNA) can be used to target virtually any genes16–18. It shows therapeutic potential for treating various diseases including cancer19–21. However, few RNAi therapeutics have entered phase II/III clinical trials22–25. This is because naked RNAs including siRNA, have poor stability in blood, don’t enter cells, and are instable in the endo/lysosomes26. Nanotechnology has demonstrated potential for overcoming the challenges facing RNAi based therapy27–31. Nanoparticles can prolong the half-life of RNAs in blood, preferentially accumulate in tumour, enhance cellular uptake, and allow for stimuli-responsive release of payload32–36. However, the RNAs released from nanoparticles could be easily degraded in endosomes/lysosomes after cellular uptake by endocytosis26,37. Therefore, it is crucial to achieve endo/lysosomal escape for effective release of siRNAs into the cytosol so that they can interact with the RNAi machinery to induce gene silencing38–41.
With the aim of enhancing bioavailability and improving endo/lysosomal escape of siRNA, we designed low pH-activated “nano-bomb” nanoparticles to deliver POLR2A siRNA (siPol2) and precisely target POLR2A in TP53loss TNBC. Carbon dioxide (CO2) can be generated from the nanoparticles under the reduced pH in endo/lysosomes to give the “nano-bomb” effect, which triggers endo/lysosomal escape for enhanced cytosolic siRNA delivery. Our data show that POLR2A suppression with the siPol2-laden nanoparticles (siPol2@NPs) leads to an enhanced reduction of the growth of POLR2Aloss tumour with no evident systemic toxicity.
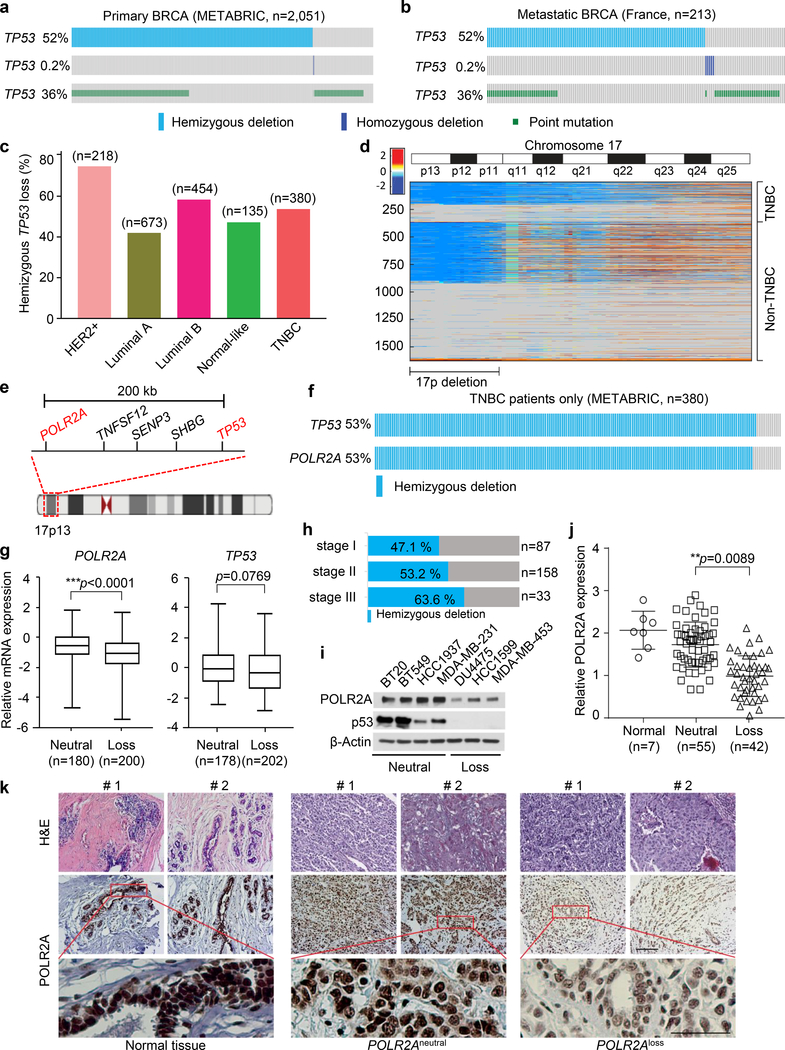
Hemizygous deletion of POLR2A and TP53 in breast cancer
Inactivation of TP53 is a frequent event in most human tumours14. However, neither TP53 mutation nor complete deletion of TP53 is the most frequent in primary human breast cancers. There are only 36% (741 out of 2,051) and 0.5% (5 out of 2,051) cases for mutation and homozygous deletion, respectively (Fig. 1a). On the contrary, hemizygous deletion of TP53 is highly frequent in both primary and metastatic breast cancers (52% and 55%, respectively, Fig. 1a, b). Particularly, 53% (202 out 380) of TNBC cases carry the hemizygous deletion of TP53 (Fig. 1c). Our analyses of the breast cancer genomes show that the TP53 deletion is within a large fragment deletion of Chr17p spanning over ~19.8 megabases of DNA in TNBC and other human breast cancers (Fig. 1d, e). The POLR2A and TP53 genes are nearly always co-deleted in the Chr17p deletion region (Fig. 1f). Importantly, the POLR2A expression levels are significantly correlated with the copy numbers of POLR2A in the TNBC subtype while this correlation is not significant for p53 (Fig. 1g and Supplementary Fig. 1). Although one allele deletion of the Chr17p fragment significantly decreases the POLR2A mRNA expression, more severe stages of TNBC are associated with increased frequencies of patients with hemizygous TP53 loss (Fig. 1h). This indicates one copy of POLR2A is sufficient to maintain cell survival. This positive correlation is validated in several TNBC cell lines (Fig. 1i). In immunohistochemical analysis using a tumour tissue microarray containing 100 TNBC samples, the expression of POLR2A was scored according to the staining intensity and proportion of signals in each sample. Accordingly, the copy numbers of POLR2A in the tumour tissue samples were determined by quantitative polymerase chain reaction (PCR). A tight correlation was validated between POLR2A copy numbers and protein expression levels (Fig 1j, k). Collectively, these data suggest inhibition of POLR2A is an amenable approach for targeted treatment of TNBC.
Fig. 1. POLR2A is almost always deleted together with TP53 in triple negative breast cancers.
a-b, Genomic alterations of TP53 (hemizygous deletion and heterozygous deletion, and point mutation) in The Cancer Genome Atlas primary (a) and metastatic (b) breast cancer dataset determined by cBioportal (n = 2,051 and 213 biologically independent samples for primary and metastatic cancer, respectively). c, The frequency of hemizygous TP53 loss in five major human breast cancer subtypes (n indicates the number of biologically independent samples for each subtype). HER2: Human epidermal growth factor receptor 2. d, Heatmap of genomic segment copy-number abnormalities (log-ratio measurements) of human chromosome 17 in triple negative breast cancers (TNBCs) as well as all the other invasive non-TNBC breast carcinomas. Positive log-ratios indicate degree of copy number gain (red) whereas negative values present the loss (blue). e, A schematic diagram showing genes adjacent to TP53 in human genome. f, Concomitant deletion of POLR2A in TNBC containing hemizygous loss of TP53. g, Correlation between gene expression and copy number variation for POLR2A and TP53 genes in breast tumours (n indicates the number of biologically independent samples). The Box-Whisker plots present a five-number summary: minima, lower quartile, centre, upper quartile, and maxima. Error bars denote mean ± s.d., and statistical significance was assessed by Student’s t-test (unpaired and two-tailed). h. The frequency of TNBC patients with hemizygous TP53 deletion at stages I, II, and III, respectively. i, Protein levels of POLR2A and p53 in seven different human TNBC cell lines (the experiments were repeated three times independently). j-k, Quantification of POLR2A expression in human breast normal, POLR2Aneutral and POLR2Aloss TNBC tumour tissue samples (j), and the representative images are shown in k. In k, two representative samples are shown for each of the three of tissues and the experiments were repeated three times independently. Error bars denote mean ± s.d., and statistical significance was assessed by Student’s t-test (unpaired, two-tailed). **: p < 0.01 and ***: p < 0.001. Scale bar, 100 μm. METABRIC and France indicate the origin of the data in The Cancer Genome Atlas (http://atlasgeneticsoncology.org/tcga_projects.html).
Synthesis and characterization of nanoparticles
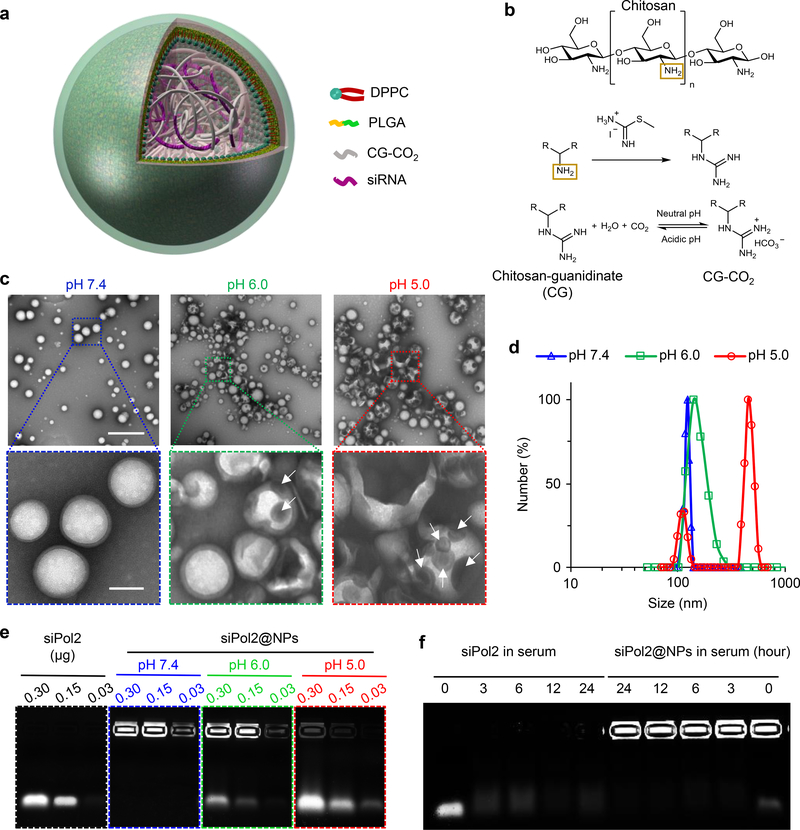
We developed a core-shell nanoplatform illustrated in Fig. 2a for delivering siPol2 to target POLR2A. The core contains siRNA and chitosan modified with guanidine (chitosan-guanidinate or CG, Fig. 2b and Supplementary Fig. 2). The guanidine group can react reversibly with CO2 to form chitosan-guanidinate-CO2 (CG-CO2) in a pH dependent manner42, which is utilized to capture/store CO2 at neutral pH for release at reduced pH such as that (~5) inside endo/lysosomes (Fig. 2b).
Fig. 2. Synthesis and characterization of nanoparticles for stabilizing POLR2A targeting siRNA.
a, A Schematic illustration of the core-shell structured nanoparticle that synthesized using the double-emulsion approach of water-in-oil-in-water (w/o/w). The aqueous phase containing POLR2A targeting siRNA (siPol2) and chitosan-guanidinate-CO2 (CG-CO2) was encapsulated in the core of the nanoparticle. Poly(lactic-co-glycolic acid) (PLGA) and dipalmitoylphosphatidylcholine (DPPC) in the organic phase (oil, dichloromethane) were used to form the shell of the nanoparticle. b, Illustrations of the chemical reactions for modification of chitosan with guanidine and for the pH dependent capture/release of CO2 with the guanidine-modified chitosan. c, Transmission electron microscopy (TEM) images of the nanoparticles under different pH conditions for 3 h at 37 °C. The nanoparticles maintain a spherical morphology and core-shell structure at pH 7.4. At pH 6.0, the nanoparticles become enlarged with a damaged shell (arrows indicated defects on the shell), probably due to the generation of CO2 inside the nanoparticles. The change is more evident at pH 5.0 with the appearance of an additional peak for the enlarged nanoparticles. Scale bars: 500 nm and 100 nm for low and high magnification images shown in the top and bottom rows, respectively. d, Nanoparticle size distribution determined by dynamic light scattering (DLS) at pH 7.4, 6.0, and 5.0. e, Electrophoretic gel assay showing the low pH-responsive release of siPol2 from siPol2@NPs in PBS (RNase free). f, Electrophoretic gel assay showing the stability of free siPol2 compared to siPol2 encapsulated in the nanoparticles (siPol2@NPs) after incubating them in serum at 37 °C for up to 24 h. The observable signal of siPol2@NPs at 24 hours indicates that the nanoparticle encapsulation could protect siPol2 from degradation in serum, while the signal of free siPol2 in serum disappeared quickly. The experiments for c-f were repeated three times independently.
Typical transmission electron microscopy images of the siPol2-laden nanoparticles (siPol2@NPs) are shown in Fig. 2c. The siPol2@NPs have a spherical morphology and core-shell structure. The nanoparticles are stable at neutral pH and 110 ± 5.7 nm in diameter with a narrow size distribution (Fig. 2c, d) and negatively charged surface (or surface zeta potential: −22.4 ± 2.1 mV, Supplementary Fig. 3). The size of the nanoparticles synthesized using CG without CO2 is not significantly affected by either pH 6.0 or pH 5.0 (Supplementary Fig. 4). In contrast, when CG-CO2 is used, their size changes to 138 ± 25.2 nm and defects (white arrows, Fig. 2c) show up on some of the nanoparticles at pH 6.0. When the pH is further reduced to 5.0, the size increases by >4 times to 463 ± 54.8 nm for most nanoparticles (resulting in an additional peak, Fig. 2d) and defects show up on all nanoparticles (Fig. 2c). These results indicate that the CO2 generation from CG-CO2 encapsulated in the nanoparticles under low pH (particularly pH 5.0) conditions could expand and/or break open the nanoparticles, to give the “nano-bomb” effect. It is also possible that the damaged nanoparticles might form aggregates at pH 5.0, which might contribute to the additional peak at ~450 nm shown in Fig. 2d.
The migration of siPol2 into electrophoretic agarose gel is almost completely inhibited by nanoparticle encapsulation with negligible release at pH 7.4 (Fig. 2e). Moreover, the strong signal observed under pH 5.0 closely resembles the free siPol2 band. Under pH 6.0, smaller bands indicating slower release of the siRNA could be seen. These data demonstrate the low pH activated “nano-bomb” effect of the nanoparticles could trigger the release of the encapsulated siPol2. Furthermore, the siPol2@NPs were observable for at least 24 h in serum while most of the free siPol2 degraded within 10 min and no siPol2 was discernible after 3 h (Fig. 2f and Supplementary Fig. 5). These data indicate the nanoparticles could enhance the stability of the siPol2 in blood and allow for pH triggered release of the siRNA.
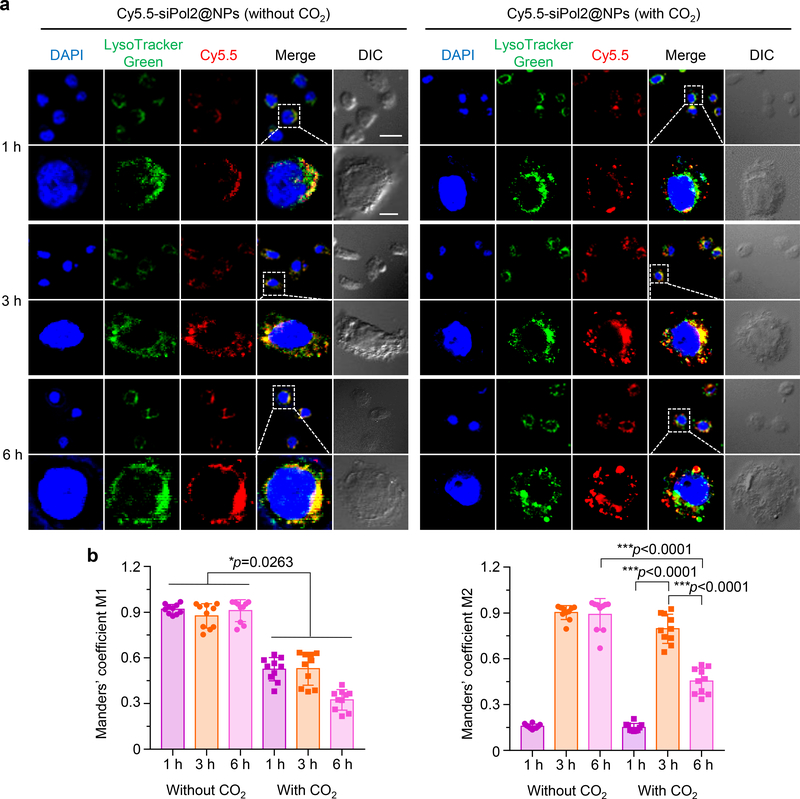
To examine cellular uptake and intracellular trafficking, siPol2 was labelled with a red fluorescence probe (Cy5.5) and encapsulated in the nanoparticles. In MDA-MB-453 TNBC cells treated using nanoparticles without CO2, the red fluorescence of Cy5.5 overlaps with the green fluorescence of LysoTracker Green that stains the endo/lysosomes at all time points (Fig. 3a, left panel). In stark contrast, the overlap between the green and red fluorescence is reduced in the cells treated using nanoparticles with CO2 at all the three time points (particularly at 6 h) (Fig. 3a, right panel), indicating successful escape of the Cy5.5-siPol2 from endo/lysosomes. These observations are confirmed by quantitative analyses of co-localization of Cy5.5-siPol2 with endo/lysosomes in the confocal fluorescence images using the Manders’ coefficients M1 and M243. As shown in Fig. 3b, M1 is close to 1 at all the three time points for the condition without CO2, indicating the siRNA encapsulated in the nanoparticles without CO2 was taken up via endocytosis and stayed in endo/lysosomes. In contrast, the M1 is much less than 1 at all the three time points (particularly 6 h) for the condition with CO2, indicating the CO2 generation in the nanoparticles can result in effective endo/lysosomal escape of the encapsulated siRNA. The M2 data show nearly all of the endo/lysosomes contained Cy5.5-siPol2 at 3 and 6 h in the absence of CO2 generation. For the condition with CO2 generation, the M2 increases first and then decreases. This is because the uptake of Cy5.5-siPol2 was faster than its endo/lysosomal escape in the first 3 h, and after that, the rate of endo/lysosomal escape of Cy5.5-siPol2 surpasses its uptake. Collectively, these data suggest the low pH triggered nano-bomb effect of the nanoparticles facilitates endo/lysosomal escape for cytosolic delivery of siRNAs.
Fig. 3. Low pH activated endo/lysosomal escape.
a, Typical confocal images of MDA-MB-453 TNBC cells incubated with Cy5.5-siPol2@NPs (with CO2) (nanoparticles encapsulated with Cy5.5 labelled siPol2 and CG-CO2) and Cy5.5-siPol2@NPs (without CO2) (nanoparticles encapsulated with Cy5.5 labelled siPol2 and CG) for 1, 3, and 6 h at 37 °C. The cell nuclei were stained using DAPI (blue), the endo/lysosomes were stained using LysoTracker Green (green), and siPol2 were labelled with Cy5.5 (red); DIC represents differential interference contrast. For the Cy5.5-siPol2@NPs (without CO2) group, the green and red fluorescence overlaps at all the time points. For the Cy5.5-siPol2@NPs (with CO2) treatment, the overlap is reduced at all the three time points and minimal at 6 h. Scale bars: 20 μm and 5 μm for low and high magnification images, respectively. The experiments were repeated three times independently. b, Quantitative analysis of co-localization of Cy5.5-siPol2 with endo/lysosomes labelled with LysoTracker Green. Manders’ Coefficient M1 denotes the fraction of Cy5.5-siPol2 overlapping with LysoTracker Green, and M2 denotes the fraction of LysoTracker Green overlapping with Cy5.5-siPol2. The coefficients are close “1” if they are highly co-localized (n= 10 images from three independent experiments). Error bars denote mean ± s.d., *: p < 0.05 and ***: p < 0.001. The statistical significance was assessed by Student t-test (paired, two tailed) and one-way ANOVA with a Dunnett’s host-hoc test for M1 and M2, respectively.
Suppression of POLR2A inhibits POLR2Aloss cancer cell growth
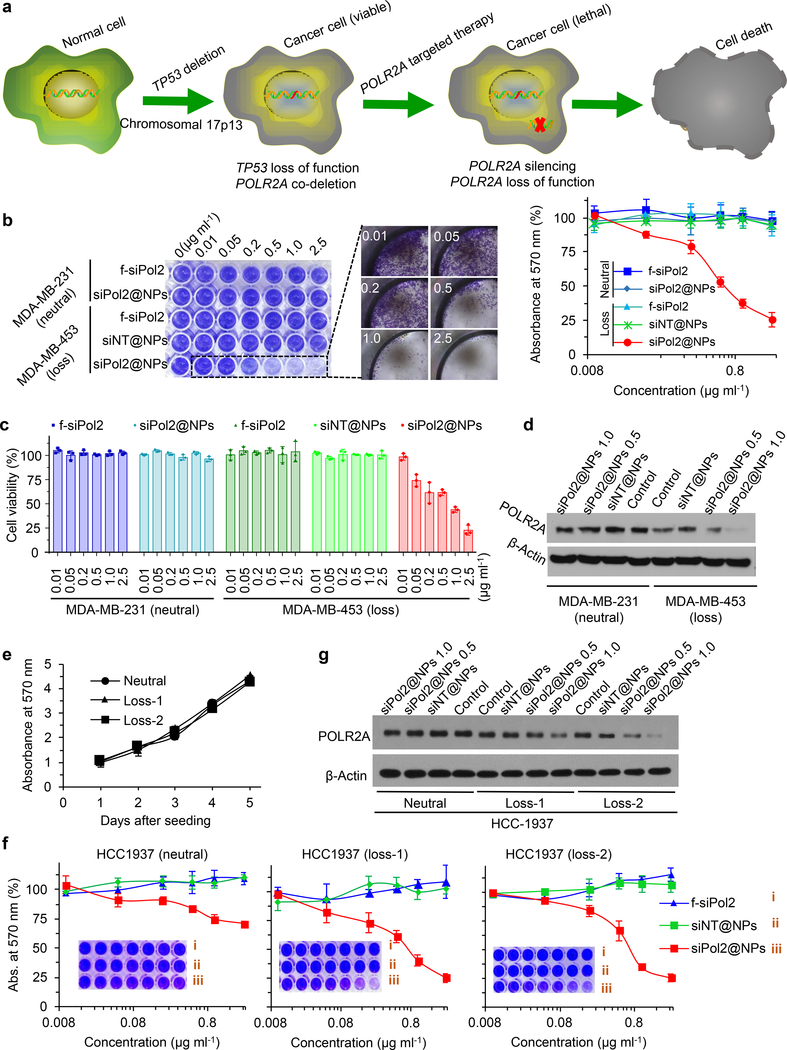
The POLR2A targeted strategy inspired by the data shown in Fig. 1 for killing TNBC cells is illustrated in Fig. 4a. To investigate the sensitivity of POLR2Aloss TNBC cells to the POLR2A inhibition, MDA-MB-453 (TP53-/mut, POLR2A−/+, or POLR2Aloss) and MDA-MB-231 (TP53+/mut, POLR2A+/+, or POLR2Aneutral) breast cancer cells were treated with siPol2@NPs together with control conditions. As shown in Fig. 4b for the colony formation assay, siNT@NPs (nanoparticles with non-targeting siRNA) is not harmful to MDA-MB-453 cells, which is expected as the nanoparticle formulation contains FDA-approved biocompatible materials. This also indicates the “nano-bomb” effect of the nanoparticles alone is not harmful to cells. The free siPol2 did not show any cell-killing effect either, due to its instability in serum and poor uptake by cells. In contrast, the siPol2@NPs could kill the MDA-MB-453 cells (POLR2Aloss) in a concentration-dependent manner although their toxicity to MDA-MB-231 cells (POLR2Aneutral) is negligible. These observations are confirmed by the cell counting kit-8 (CCK-8) assay (Fig. 4c). These results indicate the POLR2Aloss MDA-MB-453 cells are more sensitive to siPol2@NPs than POLR2Aneutral MDA-MB-231 cells, presumably because the partial loss of POLR2A renders MDA-MB-453 cells highly vulnerable to POLR2A inhibition. This is confirmed by the POLR2A protein level in the two cells after treatments (Fig. 4d). The siPol2@NPs could effectively minimize POLR2A expression in POLR2Aloss cells at a siPol2 concentration of 1.0 μg ml-1. POLR2A expression in POLR2Aneutral cells treated with the 1.0 μg ml−1 encapsulated siPol2 is still evident albeit decreased. Therefore, significant inhibition of POLR2A expression is selectively lethal to POLR2Aloss TNBC cells.
Fig. 4. Nanoparticle mediated POLR2A inhibition selectively kills POLR2Aloss cells.
a, Schematic illustration of the strategy for killing TP53loss/POLR2Aloss cells by POLR2A inhibition. POLR2A is almost always co-deleted with TP53. Moreover, POLR2A expression levels are significantly correlated to POLR2A gene copy number, inspiring the new strategy for treating TNBC. b, Wild type POLR2Aloss MDA-MB-453 (TP53-/mut, POLR2A−/+) and POLR2Aneutral MDA-MB-231 (TP53+/mut, POLR2A+/+) breast cancer cells are treated with different dosages of siPol2-laden nanoparticles (siPol2@NPs), non-target siRNA-laden nanoparticles (siNT@NPs), or free siPol2 (f-siPol2) for 72 h. Colony formation assay with crystal violet staining and quantitative analyses of absorbance at 570 nm showing the cell viability are given on the left and right, respectively (n = 3 independent experiments with 3 replicates in each experiment). c, Viability of the two types of cells quantified using the cell counting kit (CCK-8) after the aforementioned treatments (n = 3 independent experiments with 3 replicates in each experiment). d, Protein levels of POLR2A in the two types of cells without or with various treatments. Without any treatment (i.e., the two control groups), POLR2A protein expression in POLR2Aloss MDA-MB-453 cells is lower than that in POLR2Aneutral MDA-MB-231 cells. e, Cell colony assay showing that the POLR2Aloss HCC1937 (Loss-1 and Loss-2) cells have similar proliferation to the parent HCC1937 POLR2Aneutral (Neutral) cells (n = 3 independent experiments with 3 replicates in each experiment). f, Cell colony assay with Isogenic POLR2Aneutral and POLR2Aloss HCC1937 (TP53+/mut) cells for confirming the observation that POLR2Aloss cells are highly sensitive to POLR2A inhibition using siPol2@NPs. CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 system was used to generate the isogenic HCC1937 cell lines with hemizygous loss of POLR2A. siPol2@NPs could significantly inhibit the proliferation of POLR2Aloss HCC1937 (loss-1 and loss-2) cells, but not of parent POLR2Aneutral HCC1937 cells (n = 3 independent experiments with 3 replicates in each experiment). g, Protein levels of POLR2A in isogenic POLR2Aloss (Loss-1, Loss-2) and POLR2Aneutral (Neutral) HCC1937 cells without (Control) and with various treatments, showing the dose dependent inhibition of POLR2A with the siPol2@NPs. Error bars denote mean ± s.e.m. The experiments in d and g were repeated three times independently.
To exclude potential genetic differences across cell lines, we further generated two isogenic HCC1937 cell lines with hemizygous loss of POLR2A using CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (Supplementary Fig. 6a, b). The wild type HCC1937 is TP53+/mut, POLR2A+/+ (POLR2Aneutral). The parent and two isogenic POLR2Aloss HCC1937 cells exhibited similar cell growth rates (Fig. 4e), confirming hemizygous deletion of POLR2A does not affect cell survival. However, hemizygous loss of POLR2A (HCC1937loss−1and HCC1937loss−2) markedly sensitizes the HCC1937 cells to siPol2@NPs (Fig. 4f). As expected, siNT@NPs or free siPol2 had no substantial effect on the cell proliferation. POLR2A expression is reduced in isogenic HCC1937 cell lines (Fig. 4g, control groups, and Supplementary Fig. 6c). Furthermore, siPol2@NPs decreases POLR2A expression in all the cells. Collectively, these data show the materials in the nanoparticles (other than siPol2) do not have an effect on POLR2A expression in cells or their proliferation, and siPol2@NPs effectively kill POLR2Aloss isogenic TNBC cells.
Suppression of POLR2A inhibits POLR2Aloss tumour growth
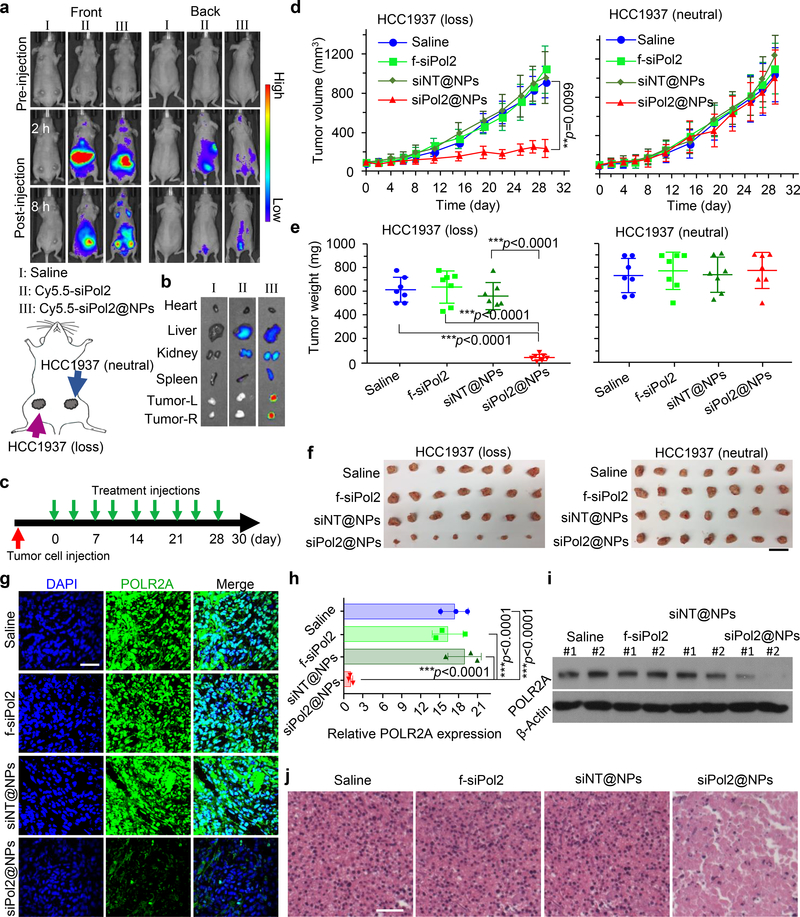
To establish orthotopic HCC1937 TNBC tumours, the isogenic POLR2Aloss and wild type POLR2Aneutral HCC1937 cells were injected into the 4th inguinal mammary fat pad on the left and right sides of each mouse (Fig. 5a). For examining the tumour targeting capability of the nanoparticles, siPol2 labelled with Cy5.5 was used to form Cy5.5-siPol2@NPs for in vivo imagining. As shown in Fig. 5a and Supplementary Fig. 7, highly enhanced fluorescence signal is observable in tumours at 8 h after administration of the Cy5.5-siPol2@NPs compared to free Cy5.5-siPol2, probably due to the enhanced permeability and retention (EPR) effect of tumour vasculature35,44. This observation is confirmed by ex vivo imaging of five major organs along with tumours harvested from the mice sacrificed at 8 h (Fig. 5b and supplementary Fig. 7). On one hand, only tumours from the Cy5.5-siPol2@NPs group present strong signal of Cy5.5, compared with tumours from the saline or free Cy5.5-siPol2 group. On the other hand, in the Cy5.5-siPol2@NPs group, the isogenic POLR2Aloss tumours on the left have similar fluorescence intensity to the wild type POLR2Aneutral tumours on the right. This indicates hemizygous loss of POLR2A does not affect the accumulation of nanoparticles in vivo, excluding the possibility that an enhanced therapeutic effect on the POLR2Aloss tumour is due to improved agent accumulation in the tumours. No accumulation of the free or encapsulated Cy5.5-siPol2 is observable in heart or kidney. Weaker fluorescence is observable in livers from the free Cy5.5-siPol2 or Cy5.5-siPol2@NPs groups. This is expected because the agents are probably cleared out of the body through liver.
Fig. 5. Targeted POLR2A inhibition selectively suppresses the growth of isogenic cells derived POLR2Aloss tumours.
a, In vivo whole animal imaging (both front and back) of Cy5.5 fluorescence at pre-injection, and 2 and 8 h after intravenous injection of saline, Cy5.5-siPol2, and Cy5.5-siPol2@NPs. The experiments were repeated three times independently. The scheme indicates the locations of tumours (the 4th inguinal mammary fat pads on both left and right sides) in mice. b, Ex vivo imaging of Cy5.5 fluorescence in tumours together with four critical organs collected after in vivo imaging at 8 h. Tumour-L and Tumour-R denote tumours on the left and right of the mouse, respectively. The experiments were repeated three times independently. c, An illustration of the treatment intervals. The mice were injected with the various treatments twice a week. d-f, Tumour growth (d), weight (e), and gross images (f) of tumours derived from isogenic POLR2Aloss and parent POLR2Aneutral HCC1937 (TP53+/mut, POLR2A+/+) cancer cells with various treatments. The data indicate that tumours with hemizygous loss of POLR2A are highly sensitive and vulnerable to further POLR2A inhibition. Error bars denote mean ± s.d. In d, the p values for comparison of siPol2@NPs versus Saline, siPol2@NPs versus f-siPol2, and siPol2@NPs versus f-siPol2 are 0.0099 (indicated on d), 0.0086, and 0.0049, respectively. Statistical significance was assessed by one-way ANOVA with a Fisher’s LSD post hoc test (d) or Dunnett’s post hoc test (e). The biologically independent sample size n=7. Scale bar: 2 cm. g-h, Immunofluorescence staining (g) of POLR2Aloss tumours and quantification of POLR2A expression (h) in tumours with various treatments (n = 3 independent experiments with 3 replicates in each experiment). The statistical significance was assessed by one-way ANOVA with a Dunnett’s post hoc test. i-j, Protein level of POLR2A (i) and H&E staining (j) of aforementioned POLR2Aloss tumours. The experiments were repeated three times independently. In i, two representative samples for each treatment are shown. Error bars denote mean ± s.d. Scale bar: 50 μm; **: p < 0.01; and ***: p < 0.001.
To investigate the material safety and antitumor efficacy, four different treatments (saline, free siPol2, siNT@NPs, and siPol2@NPs) were applied twice a week after tumour establishment in nude mice (Fig. 5c). Neither mouse death nor significant drop in body weight was observed in all the groups during the experiments (Supplementary Fig. 8a). Furthermore, no obvious damage is observable in the haematoxylin and eosin-stained histology slices of five major organs collected from mice in the siPol2@NPs and saline groups (Supplementary Fig. 8b). Moreover, no obvious change in aspartate aminotransferase and alanine aminotransferase levels is observable in wild type C57BL/6J mice with intact immune system after treated with the various formulations (Supplementary Fig. 9), indicating that the nanoparticles are well tolerated by the animal. We further measured the serum level of tumour necrosis factor-α, Interferon-γ, Interleukin-6, Interleukin-10, and monocyte chemoattractant protein-1 in the wild type mice. Although significantly increased levels of Interferon-γ and monocyte chemoattractant protein-1 were observed for the siNT@NP and siPol2@NP treatments at 6 h after injection, they returned to the baseline levels on day 1 and thereafter. No significant difference was observed between the various conditions for all the other cytokines in 7 days, indicating negligible immune response induction by the nanoparticles.
Inhibiting POLR2A by siPol2@NPs significantly inhibited the POLR2Aloss tumour growth by ~80% (left panels in Fig. 5d-f) compared with the tumour from control (saline) group. By contrast, no significant antitumor effect was observed on the POLR2Aneutral tumours (right panels in Fig. 5d-f). As expected, the POLR2A protein levels in siPol2@NPs treatment group were minimized (Fig. 5g-i). These data confirm that POLR2A silencing leads to selective suppression of POLR2Aloss tumours. Moreover, histological staining reveals extensive necrosis in the POLR2Aloss tumours only from the siPol2@NPs group (Fig. 5j).
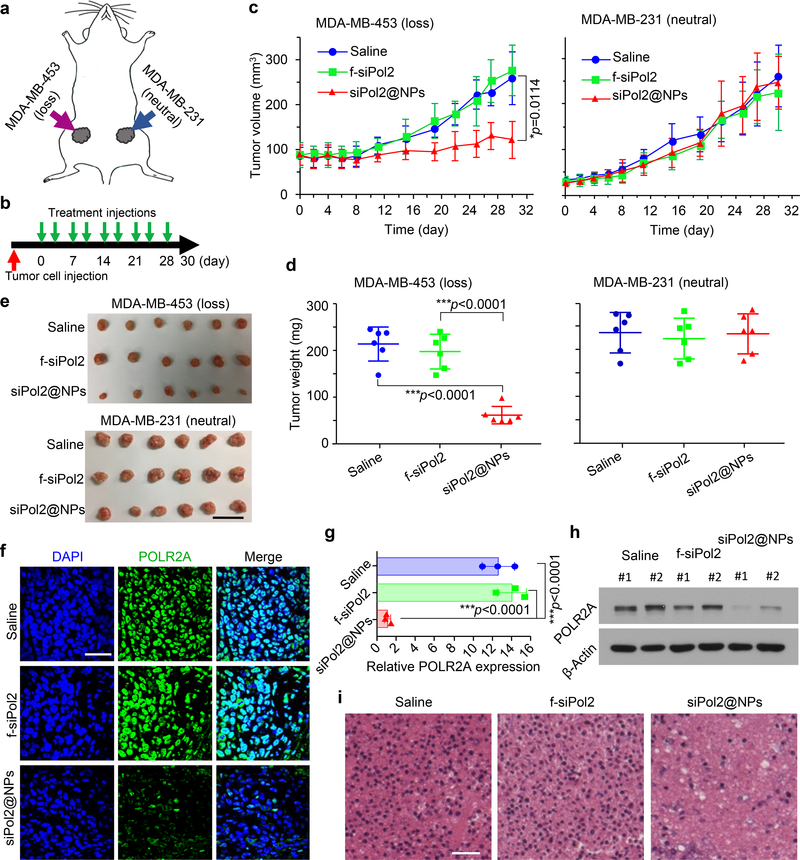
To confirm the findings with tumours derived from the isogenic pairs of HCC1937 cells, we used wild type POLR2Aloss (MDA-MB-453) and POLR2Aneutral (MDA-MB-231) cells to establish the orthotopic tumours and did treatment injections of saline, siNT@NPs, and siPol2@NPs twice a week (Fig. 6a, b). Neither mouse death nor significant body weight drop was observed in all the groups (Supplementary Fig. 10a), and no evident damage could be observed in all the five organs from in the siPol2@NPs treatment group (Supplementary Fig. 10b). Furthermore, siPol2@NPs exhibit excellent antitumor capacity to MDA-MB-453 (POLR2Aloss) tumours, while no significant difference in the growth of the MDA-MB-231 (POLR2Aneutral) tumours is observable for the three different groups (Fig. 6c-e). The siPol2@NPs treatment minimizes the POLR2A protein levels in the MDA-MB-453 tumours (Fig. 6f-h), confirming that POLR2A silencing leads to tumour suppression in the POLR2Aloss tumour model. Histological staining shows extensive necrosis in the POLR2Aloss tumours only from the siPol2@NPs group (Fig. 6l).
Fig. 6. Targeted POLR2A inhibition selectively suppresses the growth of wild type cells derived POLR2Aloss tumours.
a, A schematic illustration of the tumours established by implanting MDA-MB-453 (TP53mut, POLR2Aloss) cells on the left and MDA-MB-231 (TP53mut, POLR2Aneutral) cells on the right 4th inguinal mammary fat pads. b, An illustration of the treatment intervals. The mice were injected with the various treatments twice a week. c-e, Growth curves (c), weight (d), and gross images (e) of tumours derived from POLR2Aloss and POLR2Aneutral human TNBC cells with various treatments. The data indicate that tumours with POLR2Aloss are highly sensitive and vulnerable to further POLR2A inhibition. Error bars denote mean ± s.d. In c, the p values for comparisons of siPol2@NPs versus Saline and siPol2@NPs versus f-siPol2 are 0.0114 (indicated in c) and 0.0061, respectively. The statistical significance was assessed by one-way ANOVA with a Fisher’s LSD post hoc test for c or Dunnett’s post hoc test for d. The biologically independent sample size n=6. Scale bar: 2 cm. f-g, Immunofluorescence staining (f) of POLR2Aloss tumours and quantitative data of POLR2A expression (g) in tumours with various treatments (n=3 independent experiments with 3 replicates in each experiment). The statistical significance was assessed by one-way ANOVA with a Dunnett’s post hoc test. h-i, Protein level of POLR2A (h) and H&E staining (i) of the aforementioned POLR2Aloss tumours. The experiments were repeated three times independently. In h, two representative samples for each treatment are shown. Scale bar: 50 μm. Error bars denote mean ± s.d.; *: p < 0.05; and ***, p < 0.001.
It is worth noting that the strategy of targeting POLR2A using our siPol2@NPs can also be used to selectively inhibit the growth of POLR2Aloss HER2+ HER18 breast cancer cells (in vitro) and tumours (in vivo) (Supplementary Note 1 and Supplementary Figs. 11–12). Altogether, we show the siPol2@NPs based POLR2A suppression inhibits the growth of POLR2Aloss MDA-MB-453 (TP53-/mut, POLR2A−/+), isogenic HCC1937 (TP53mut/+, POLR2A+/−), and isogenic HER18 (TP53+/+, POLR2A+/− and TP53+/−, POLR2A+/−) cells and tumours, regardless of their TP53 status. While TP53 and POLR2A are often co-deleted in a majority of human cancers with hemizygous loss of TP53, the status of TP53 has no significant impact on the anti-cancer activity of POLR2A inhibition.
Conclusions
In this study, by analysing TNBC databases, we reveal that POLR2A gene is almost always co-deleted with TP53 in the Chr17p deletion region, and 53% of TNBC harbours this heterozygous deletion. Moreover, the POLR2A expression levels are highly correlated with the copy number of POLR2A, rendering cancer cells with heterozygous loss of TP53 vulnerable to POLR2A inhibition. This collateral loss of POLR2A with TP53 prompted us to use RNAi to precisely target POLR2A for TNBC treatment. To overcome the hurdles to cytosolic delivery of siRNA for RNAi, we developed a unique nanoplatform with a low pH-activated “bomb-like” effect for endo/lysosomal escape. This improves cytosolic delivery of siPol2 to inhibit POLR2A in TP53loss cells. The anticancer capability of siPol2@NPs in vivo was examined using three different orthotopic tumour models derived from both wild type and isogenic cell lines. The nanoparticles were found to preferentially accumulate in both the POLR2Aneutral and POLR2Aloss tumours. Importantly, our data show that, in tumours with heterozygous POLR2A loss, inhibition of POLR2A with siPol2-laden nanoparticles leads to an enhanced reduction of the tumour growth with no evident systemic toxicity. This approach also possesses several important advantages for clinical applications (Supplementary Note 2). Collectively, this study may provide a promising nanotechnology-based precision-targeting strategy for fighting against TNBC and potentially many other types of cancers harbouring the common TP53 genomic alteration, regardless of the TP53 status.
Methods
The Cancer Genome Atlas analysis.
The Cancer Genome Atlas primary (origin: METABRIC Nature 2012 & Nat Commun 2016, and Cell 2015) and metastatic (origin: France 2016) breast cancer data were downloaded from cBioPortal (http://atlasgeneticsoncology.org/tcga_projects.html), which included copy number variation at segment levels in log-ratio, copy number variation at gene levels estimated by using the GISTIC2 algorithm, RNA-seq for gene expression in base-2 log scale, and patient information on oestrogen receptor, progesterone receptor, and HER-2/neu status. We analysed the correlation between gene copy number and the corresponding gene expression as previously described14. The triple-negative breast cancer (TNBC) subtypes, defined by PAM50 profiling test, included all the basal-like and claudin-low groups.
Materials.
PLGA (lactide:glycolide: 75:25, Mw: 4,000–15,000 Da), and organic solvents were purchased from Sigma (St. Louis, MO, USA). Agarose, ethidium bromide and loading buffer were purchased from Thermo Fisher Scientific (Grand Island, NY, USA). DPPC was purchased from Anatrace (Maumee, OH, USA). Chitosan oligosaccharide of pharmaceutical grade (Mw: 1.2 kDa, 95% deacetylation) was purchased from Zhejiang Golden Schell Biochemical Co. Ltd (Zhejiang, China). Methyl aminomethanimidothioate hydroiodide was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-POLR2A antibody (sc-47701, dilution of 1:10000 for western blot and 1:200 for immunofluorescence staining), HRP-anti-rabbit IgG (sc-2030, dilution of 1:5000), HRP-anti-mouse IgG (sc-516102, dilution of 1:5000) antibodies were purchased from Santa Cruz (Dallas, TX, USA). Anti-β-actin (AM1829B, dilution of 1:5000) antibody was purchased from Abgent (San Diego, CA, USA). Alexa Fluor 488-labeled anti-mouse IgG (A11001, dilution of 1:250) antibody was purchased from Life Technologies (Waltham, MA, USA). Cell counting kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies (Rockville, MD, USA). Mouse Inflammation Kit (#552364) was purchased from BD Biosciences (San Jose, CA, USA). Alanine Transaminase (ALT) Activity Assay Kit (ab105134) and Aspartate Aminotransferase (AST) Activity Assay Kit (ab105135) were purchased from Abcam (Cambridge, MA, USA).
Nanoparticle synthesis.
The double-emulsion method45 with slight modification was used to synthesize nanoparticles in this study. First, the chitosan-guanidinate (CG) was synthesized by modifying chitosan with the guanidine group (Fig. 2b). The guanidine group is a common functional group in many natural products including the naturally occurring amino acid L-arginine. Briefly, 480 mg of chitosan (1 kDa) was dissolved in 12 ml of deionised (DI) water. A total of 6 ml of methyl aminomethanimidothioate hydroiodide in acetonitrile (145 mg ml−1) was then added and stirred at room temperature for 24 h under N2 atmosphere. Afterward, the solvent and side-product were removed under vacuum. Lastly, to remove the unreacted methyl aminomethanimidothioate hydroiodide, the solid was dissolved in 1 ml of DI water and dropped into 10 ml of tetrahydrofuran (THF), filtered, and the final solid CG was collected for further use. For capturing CO2, CG-CO2 was made by bubbling the aqueous solution of CG for 1 h with CO2. To synthesize nanoparticles, 10 mg of DPPC and 20 mg of PLGA were dissolved in dichloromethane (DCM, 2 ml). This organic solution and 400 μl of DI water dissolved with 1 mg ml−1 CG-CO2 and 500 μg ml−1 POLR2A siRNA (without or with Cy5.5 modification, Sigma) were transferred into a centrifuge tube (50 ml). The sample was then emulsified for 1 min by sonication with a Branson 450 sonifier at 18% power. A total of 8 ml of PVA solution (2% in DI water) was then added into the centrifuge tube and the sample was further emulsified as aforementioned for 1 min. Afterward, the organic solvent (i.e., DCM) was removed by rotary evaporation. The nanoparticles were then collected by centrifugation for 10 min at 10000g and washed two times in DI water at room temperature.
Nanoparticle characterization.
Both dynamic light scattering (DLS) and transmission electron microscopy (TEM) were used to characterize the nanoparticles. The nanoparticles were soaked for 6 h in either acetate buffer (pH 5.0) or phosphate buffer (for pH 6.0 and pH 7.4). For TEM study, uranyl acetate solution (2%, w/w) was used to negatively stain the nanoparticles before examining them with the Tecnai G2 Spirit transmission electron microscope from FEI (Moorestown, NJ, USA). The size distribution of the nanoparticles was studied using a 90 Plus/BI-MAS DLS instrument from Brookhaven (Holtsville, NY, USA). The encapsulation efficiency of the siRNA was 68.6 ± 7.2%, which was calculated as the ratio of the amount of the siRNA encapsulated in the nanoparticles to that the total amount of the siRNA fed for encapsulation. The loading content of the siRNA in the nanoparticles was 0.73 ± 0.25%, which calculated as the ratio of the amount of the siRNA encapsulated in the nanoparticles to that the total amount of the nanoparticles including the siRNA. Both the encapsulation efficiency and loading content were quantified by using Cy5.5-siPol2 for encapsulation. The amount of Cy5.5-siPol2 in a sample was measured spectrophotometrically using a Beckman Coulter (Indianapolis, IN, USA) DU 800 UV-vis Spectrophotometer based on its absorbance peak at 670 nm. A standard curve of free Cy5.5-siPol2 (absorbance vs. concentration) was used for converting the measured absorbance into the concentration of Cy5.5-siPol2 in a sample.
Electrophoretic gel assay.
Free POLR2A siRNA (siPol2) and siPol2@NPs (in phosphate buffered saline or serum) were mixed with loading buffer, and then loaded into 2%wt agarose gel with 0.5 mg ml−1 ethidium bromide. Electrophoresis was conducted in 1x tris-acetate-EDTA (TAE) buffer at 80 V for 10 min. The resulting gels were analysed using a UV illuminator (FluorChemTM E System, CA, USA) to show the location of siPol2. Uncropped scans of the gels are presented in the Supplementary Fig. 13.
Cell culture.
MDA-MB-231, MDA-MB-453, and HCC1937 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured under the standard conditions specified by ATCC. HER18 cells (stably overexpress HER2) were a gift from Dr. Mong-Hong Lee (MD Anderson Cancer Centre). The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum at 37 °C in 5% CO2. Cell identity was confirmed by validating the STR DNA fingerprinting using the AmpFLSTR Identifiler Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
Generation of POLR2A-heterozygous cell lines.
The generation of POLR2Aloss isogenic cell lines was conducted using CRISPR/Cas9 as described previously14. Briefly, 2 × 106 of HCC1937 or HER18 cells were transfected with 2 μg of Cas9/sgRNA-expressing vector DNA using a Nucleofector kit (Lonza, Walkersville, MD, USA). Genome editing efficacy was tested by the Surveyor assay. Single colonies were isolated and the PCR products from positive clones were ligated to the pGEM-T Easy Vector (Promega, Madison, WI, USA) and further confirmed by Sanger DNA sequencing.
Cellular uptake and intracellular distribution.
The nanoparticles were encapsulated with Cy5.5-siPol2 using the aforementioned double-emulsion method. For the nanoparticles without CO2, CG was used instead of CG-CO2. MDA-MB-453 cells were then treated with the Cy5.5-siPol2@NPs at 37 °C for up to 6 h. Afterward, the cells were incubated at 37 °C for 30 min with 50 nM DAPI and 90 nM LysoTracker Green in cell culture medium. The cells were then fixed with 4% paraformaldehyde and examined using a FluoView FV1000 confocal microscope from Olympus (Centre Valley, PA, USA). For quantitative analysis of the co-localization between Cy5.5-siPol2 and LysoTracker Green inside cells, the Manders’ co-localization coefficients between the fluorescence signals of Cy5.5-siPol2 and LysoTracker Green were calculated using ImageJ (ImageJ bundled with 64-bit Jave 1.8.0_112, https://imagej.nih.gov/ij) with the JACoP co-localization plugin module. The two Manders’ coefficients M1 and M2 were calculated as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where the subscript i represents the ith pixel in the fluorescence image, R represents red fluorescence intensity, G represents green fluorescence intensity, T1 represents the threshold for red channel, and T2 represents the threshold for green channel. The two fluorescence intensities and thresholds were determined by the built-in algorithm of the JACoP co-localization plugin module of ImageJ for both the green and red channels.
Immunoblotting.
Immunoblotting was performed as described previously14. Briefly, cell lysates were made in lysis buffer (pH 7.5) containing 1 mM EDTA, 50 mM Tris, 150 mM NaCl, 0.5% Triton X-100, 0.5% NP-40, 1 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 5 mM sodium vanadate, and 1 μg/ml leupeptin, aprotinin, and pepstatin. Proteins were then separated by SDS-PAGE gels and further transferred to the membranes of polyvinylidene difluoride (Bio-Rad, Hercules, CA, USA). After blocking the membranes with 5% nonfat milk for 1 h at room temperature, they were incubated with primary antibodies as indicated. The membranes were then washed and incubated at room temperature with peroxidase-conjugated secondary antibodies for 1 h. After 5 times of washing, immunodetected bands on the membranes were visualized by taking chemiluminescent images on X-ray films with the enhanced chemiluminescence (ECL) system (PerkinElmer, Waltham, MA, USA) as per the manufacturer’s instructions. All uncropped scans are shown in Supplementary Fig. 13 for the western blots.
Cell viability and cell colony formation assays.
Equal numbers of cells were plated in 96 well plates in triplicate. After incubation with indicated treatments for 72 hours, cell viability was quantified using the CCK-8 according to the manufacturer’s instructions. For the cell colony formation assay, cell colonies were visualized using crystal violet (Fisher scientific, Pittsburgh, PA, USA) according to the manufacturer’s instructions.
Animals.
Female NU/NU nude mice (6 weeks old) were purchased from either Charles River (Wilmington, MA, USA) for the TNBC tumour models or The Jackson Laboratory (Bar Harbour, ME, USA) for the HER2+ tumour model. Wild type C57BL/6J mice (6 weeks old) were purchased from The Jackson Laboratory. All animals were maintained 16:8 h of light-dark cycle. All animal studies were conducted by following protocols approved by the Institutional Animal Care and Use Committee (IACUC) at The Ohio State University and Indiana University School of Medicine. The animal protocols are compliant with all relevant ethical regulations.
Orthotopic xenograft breast tumour models and treatments.
For wild type POLR2Aloss/POLR2Aneutral orthotopic xenograft TNBC model, nude mice were injected with 1 × 106 MDA-MB-453 cancer cells into the 4th inguinal mammary fat pad on the left and 1 × 106 MDA-MB-231 cancer cells into the 4th inguinal mammary fat pad on the right on the same day. For isogenic POLR2Aloss/POLR2Aneutral orthotopic xenograft TNBC model, nude mice were injected with 1 × 106 HCC1937 (POLR2Aloss) cancer cells into the 4th inguinal mammary fat pad on the left and HCC1937 (POLR2Aneutral) cancer cells into the 4th inguinal mammary fat pad on the right on the same day. For isogenic POLR2Aloss/POLR2Aneutral orthotopic xenograft HER2+ breast cancer model, nude mice were injected with 5 × 106 HER18 (POLR2Aloss) cancer cells into the 4th inguinal mammary fat pad on the left and HER18 (POLR2Aneutral) cancer cells into the 4th inguinal mammary fat pad on the right. The nude mice were supplemented with weekly subcutaneous oestradiol cypionate injections (3 mg kg−1 per week), starting 1 week prior to injection of tumour cells. After initial tumour establishment (~100 mm3), mice were randomly grouped and treated with indicated formulations. Treatment formulations were administered twice weekly by intravenous injection via the tail vein, and tumour size and body weight were monitored biweekly. The dose of siPol2 for any formulations with the siRNA was 0.3 mg kg−1 body weight. Tumour size was measured using a calliper, and tumour volume was calculated using the standard formula: 0.5 × L × W2, where L is the long diameter and W is the short diameter. Mice were euthanized when they met the institutional euthanasia criteria for tumour size and overall health condition. Tumours were removed, photographed, and weighed. The freshly dissected tumour tissues were either used for western blot analysis or fixed in 10% buffered formalin overnight, transferred to 70% ethanol, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) and indicated antibodies.
Biodistribution of nanoparticles in vivo.
After initial tumour establishment (~100 mm3), mice were injected with either 100 μl of saline, Cy5.5-siPol2 (50 μg) in 100 μl of saline, or Cy5.5-siPol2@NPs (nanoparticles containing 50 μg of Cy5.5-siPol2) in 100 μl of saline. Images were taken immediately before injection and at 2 and 8 h after intravenous injection via the tail vein using an in vivo imaging system (Perkin Elmer IVIS, Waltham, MA, USA) with an excitation at 675 nm and a 690–770 nm Cy5.5-filter to collect the fluorescence emission of Cy5.5. For ex vivo imaging, the mice were sacrificed after in vivo imaging at 8 h, and tumor and main organs were harvested for further fluorescence imaging using the same in vivo imaging system.
Detection of immune response induction by nanoparticles.
Wild type C57BL/6J mice (6 weeks old) purchased from The Jackson Laboratory were randomized and injected intravenously with the following treatments: 1) saline, 2) free siPol2, 3) siNT@NPs, and 4) siPol2@NPs. At indicated time points, blood was drawn from mice and the concentration of indicated cytokines in serum was measured by BD Mouse Inflammation Kit (#552364), ALT Activity Assay Kit (Abcam, ab105134), and AST Activity Assay Kit (Abcam, ab105135) according to the manufacturer’s instructions.
Statistics and reproducibility.
Each experiment was repeated independently for at least three times. Unless otherwise noted, data are presented as mean ± standard deviation (s.d.) or standard error of mean (s.e.m.). Student’s t-test (unpaired and two-tailed) was used to compare two groups of independent samples, assuming equal variance with no samples being excluded from the analysis. One-way analysis of variance (ANOVA) with Dunnett’s post hoc analysis was used for multiple comparison (when more than two groups compared). One-way ANOVA with Fisher’s LSD post hoc was used for tumour growth analysis. Statistical methods used for The Cancer Genome Atlas data analysis were described above. All statistical analyses were carried out with Prism (version 7.0, GraphPad Software, San Diego, CA, USA). A p value less than 0.05 was considered statistically significant.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
This work was partially supported by grants from American Cancer Society (ACS #120936-RSG-11–109-01-CDD) and NIH (R01CA206366) to X.H. and X.L., Vera Bradley Foundation for Breast Cancer Research to X. L., Pelotonia post-doctoral Fellowship to J.X., and American Cancer Society Institutional Research Grant to Y.L.
Footnotes
Competing interests
X.H. and X.L. have applied for patents related to this study.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permission information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to X.H or X.L.
References
- 1.Dent R, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res 13(15), 4429–4434 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE, Reis-Filho JS Triple-Negative Breast Cancer. N. Engl. J. Med 363(20), 1938–1948 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486(7403), 395–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denkert C, Liedtke C, Tutt A, von Minckwitz G Molecular alterations in triple-negative breast cancer—the road to new treatment strategies. Lancet 389(10087), 2430–2442 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Mayer EL, Burstein HJ Chemotherapy for Triple-Negative Breast Cancer: Is More Better? J. Clin. Oncol 34(28), 3369–3371 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Chin L, Hahn WC, Getz G, Meyerson M Making sense of cancer genomic data. Gene Dev 25(6), 534–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, et al. Cancer Genome Landscapes. Science 339(6127), 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18(1), 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol 13(11), 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisman PS, et al. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod. Pathol 29, 476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445(7128), 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Olivier M, et al. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum. Mutat 19(6), 607–614 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Joerger AC, Fersht AR The p53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem 85(1), 375–404 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature 520(7549), 697–701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark VE, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat. Genet 48, 1253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novina CD, Sharp PA The RNAi revolution. Nature 430(6996), 161 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Liang C, et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference–based bone anabolic strategy. Nat. Med 21, 288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ Drugging the undruggable RAS: Mission Possible? Nat. Rev. Drug Discov 13, 828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul CP, Good PD, Winer I, Engelke DR Effective expression of small interfering RNA in human cells. Nat. Biotechnol 20, 505 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Morris KV, Chan SW-L, Jacobsen SE, Looney DJ Small Interfering RNA-Induced Transcriptional Gene Silencing in Human Cells. Science 305(5688), 1289–1292 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature 448, 39 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Wittrup A, Lieberman J Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bobbin ML, Rossi JJ RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol 56(1), 103–122 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Dahlman JE, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol 9, 648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuckerman JE, Davis ME Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov 14(12), 843–856 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Wang H, et al. A near‐infrared laser‐activated “nanobomb” for breaking the barriers to microRNA delivery. Adv. Mater 28(2), 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Cui J, et al. Ex vivo pretreatment of human vessels with siRNA nanoparticles provides protein silencing in endothelial cells. Nat. Commun 8(1), 191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol 7(6), 389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong HJ, Mooney DJ Microenvironmental regulation of biomacromolecular therapies. Nat. Rev. Drug. Discov 6(6), 455–463 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Shu D, et al. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 9(10), 9731–9740 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams BD, et al. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer Res 76(4), 927–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X, Huang L Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res 45(7), 971–979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater 11, 82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Yu J, Lu X, He X Nanoparticle systems reduce systemic toxicity in cancer treatment. Nanomedicine (Lond) 11(2), 103–106 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Farokhzad OC, Langer R Impact of nanotechnology on drug delivery. ACS Nano 3(1), 16–20 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Aimetti AA, Langer R, Gu Z Bioresponsive materials. Nat. Rev. Mater 2, 16075 (2016). [Google Scholar]
- 37.Kim HJ, Kim A, Miyata K, Kataoka K Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug. Deliv. Rev 104, 61–77 (2016). [DOI] [PubMed] [Google Scholar]
- 38.El Andaloussi S, Mäger I, Breakefield XO, Wood MJA Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov 12, 347 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Maas SLN, Breakefield XO, Weaver AM Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 27(3), 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol 29, 341 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, et al. Polymetformin combines carrier and anticancer activities for in vivo siRNA delivery. Nat. Commun 7, 11822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seipp CA, Williams NJ, Kidder MK, Custelcean R CO2 capture from ambient air by crystallization with a guanidine sorbent. Angew. Chem. Int. Ed. Engl 56(4), 1042–1045 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Manders EMM, Verbeek FJ, Aten JA Measurement of co-localization of objects in dual-colour confocal images. J. Microsc 169(3), 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater 1, 16014 (2016). [Google Scholar]
- 45.Wang H, et al. Multi-layered polymeric nanoparticles for pH-responsive and sequenced release of theranostic agents. Chem. Commun. (Camb) 51(36), 7733–7736 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding authors upon request.