Figure 4 – AcrVA1-triggered endoribonuclease activity truncates a Cas12a-bound crRNA.
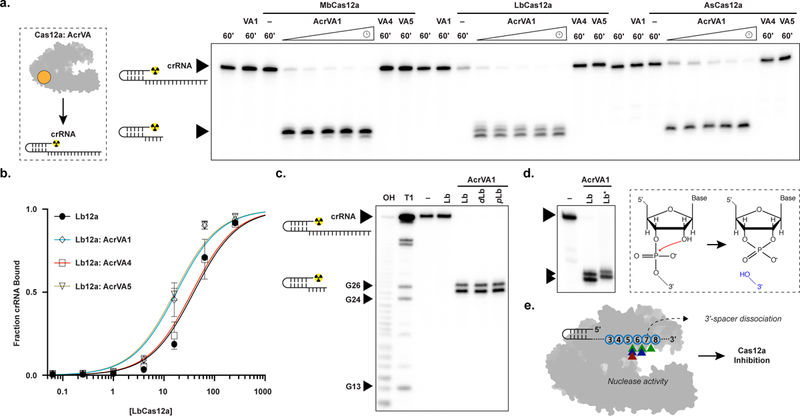
a) Radiolabeled kinetic crRNA cleavage assays for (left to right) MbCas12a, LbCas12a, and AsCas12a complexed with or without AcrVAs. Time courses represent 1, 2, 5, 15, and 60 min. Black triangles indicate full-length and truncated crRNA, b) Quantified fraction crRNA bound by LbCas12a in the presence or absence of AcrVAs determined by EMSA (mean ± sd, n = 3 independent experiments). Measured dissociation constants (Kd) are 38.9 nM ± 4.7, 17.6 ± 2.4, 35.8 ± 4.4, and 16.4 ± 2.1 in absence of inhibitor or in the presence of AcrVA1, AcrVA4, or AcrVA5, respectively, Source data are available with the paper online. c) Radiolabeled crRNA cleavage assay with LbCas12a-crRNA complexed without or with AcrVA1. Treatments in the absence of AcrVA1 are (left to right) crRNA hydrolysis ladder (OH), crRNA RNase T1 digestion (T1), untreated crRNA (−), and crRNA incubated with LbCas12a (Lb). Treatments in the presence of AcrVA1 are (left to right) wild-type LbCas12a (Lb), dLbCas12a (D832A, dLb), and processing dead Cas12a (K785A, pLb). A large black triangle indicates the full-length crRNA, smaller triangles indicate RNase T1 mapped G-nucleotides, d) Radiolabeled crRNA cleavage assay using LbCas12a-crRNA complexed with AcrVA1 that is either untreated (Lb) or treated with PNK (Lb*), e) Schematic representation of AcrVA1-triggered crRNA spacer cleavage activity on Cas12a. Cleavage sites for Mb (blue), Lb (green), and AsCas12a (red) are indicated with triangles. The uncropped gel images are available in Supplementary Data Set 1.
