Abstract
Background
Tobacco use is responsible for approximately 80–90% of non‐small cell lung cancer cases. A large evidence base has shown that the ERBB pathway is associated with the occurrence of lung cancer. However, the mechanisms of how smoking activates the ERBB pathway have yet to be explained. We hypothesized that microRNAs may induce ERBB pathway activity during the process of lung cancer carcinogenesis.
Methods
We analyzed microRNA array data from the Gene Expression Omnibus and the Kyoto Encyclopedia of Genes and Genomes to determine any associations between genes and smoking in three groups of patients with NSCLC: smokers, former smokers, and non‐smokers.
Results
The interaction network among miRNAs, including hsa‐mir‐185‐3p, hsa‐mir‐4295, hsa‐mir‐4288, and hsa‐mir‐613, promotes lung cancer development by affecting the ERBB pathway.
Conclusion
Our findings provide evidence to explain the mechanism of lung cancer development in smokers.
Keywords: EGFR, lung cancer, microRNA, smoking
Introduction
Non‐small cell lung cancer (NSCLC) is the leading cause of tumor death and the most common malignant disease worldwide.1 Smoking is the main contributor to lung cancer development, responsible for approximately 90% of cases.2 Tobacco smoke contains more than 60 cancer‐causing agents that can induce DNA damage and mutation in human cells.3 It induces activation of multiple regulatory pathways, including Wnt/β‐catenin, PKA‐CREB, and ERBB pathways. EGFR mutations or amplification have been observed in normal bronchi,4 normal bronchial epithelium, hyperplastic bronchial epithelium,5 and atypical adenomatous hyperplasia6 of lung cancer patients with EGFR mutations, suggesting that these changes are early events in the pathogenesis of NSCLC. Therefore EGFR‐related pathway activation is regarded as the main cause of lung carcinogenesis in smokers; however, the mechanisms have not yet been fully explained.
The EGFR super family, a widely expressed cell surface protein family, is thought to participate in cancer development and progression.7 EGFR is now used to assist in the diagnosis of lung cancer and is a target of anticancer drug treatment.8 The EGFR family consists of four members: EGFR (ERBB1, HER1), ERBB2 (HER2), ERBB3 (HER3), and ERBB4 (HER4). In addition to the formation of homologous dimers after ligand binding, EGFR can also form an allogeneic dimer with another member of the family, such as HER2, which stabilizes ligand binding and enhances activation of the downstream signal pathway.9 Overexpression of HER2 occurs in 32% of NSCLC patients, and in 2–23% of cases, this is the result of an increase in the number of gene copies; patients with HER2 overexpression have relatively short survival.10 Many mouse models have been established to study the role of the EGFR family in the development of lung cancer. EGFR mutant transgenic mice show typical adenomatous hyperplasia at seven weeks old and adenocarcinoma at four weeks, accompanied by high expression of HER2 and ERBB3. Treatment with gefitinib (an EGFR‐tyrosine kinase inhibitor) can effectively inhibit the growth of tumors harboring EGFR mutations, without lethal toxicity.11 Thus, EGFR promotes cell proliferation, activates the ERBB pathway, and induces carcinogenesis. However, how tobacco use induces upregulation of ERBB pathway‐related genes has not been determined.
MicroRNAs (miRNAs) are small, noncoding RNA molecules (containing approximately 22 nucleotides) found in plants, animals, and some viruses, which act in the RNA silencing and posttranscriptional regulation of gene expression.12 Changes in miRNA expression can lead to tumor transformation.13 IGBP1 is commonly expressed in lung adenocarcinoma, but particularly in the early stage. MiR‐3941 is a tumor suppressor miRNA that directly inhibits and regulates IGBP1. Overexpression of miR‐3941 and inhibition of IGBP1 induce apoptosis by increasing the rate of cleavage of Caspase‐3 and poly (ADP‐ribose) polymerase.14 MiRNA‐125b is also involved in early changes of tumor suppressing miRNAs in prostate cancer. There are many miRNAs that regulate cancer cell proliferation by the ERBB pathway in lung cancer. MicroRNA‐145 inhibits migration and induces apoptosis in human NSCLC cells by regulating the EGFR/PI3K/AKT signaling pathway.15 MicroRNA‐133a downregulates EGFR expression in human NSCLC cells via AKT/ERK signaling.16 MicroRNA‐30b inhibits NSCLC cell growth by targeting EGFR.17 MiR‐125b is directly targeted to ERBB2/B3 and MET, and the absence of miR‐125b leads to enhanced signals by the Met regulated PI3K/AKT and Ras/PMEK pathways.18 These results show that different miRNAs affect cell proliferation and invasion by the same ERBB pathway; however whether miRNAs can regulate the development of lung cancer via the ERBB pathway in smokers is not yet known.
In this study, gene expression data from smokers with and without lung cancer were analyzed using a systems biology approach that included Gene Oncology and enrichment analysis of differentially expressed genes between normal and cancerous lungs to identify the potential key factors contributing to lung cancer progression. We found comprehensive changes in microRNA expression. Moreover, hsa‐mir‐185‐3p, hsa‐mir‐4295, hsa‐mir‐4288, hsa‐mir‐613, and other genes can regulate the downstream proteins of the EGFR pathway through the regulation of target genes. Our findings suggest the possible mechanism of lung carcinogenesis in smokers.
Methods
Target predictions of lung cancer‐related microRNAs (miRNAs)
TargetScan (http://www.targetscan.org) was used to generate lists of possible gene targets of each miRNA. The targeted genes were input into another web server, Panther (http://www.pantherdb.org/), which is designed for gene function clusters. Panther analysis provided the protein classes and we clustered the same functional classes of proteins with the top 10 classes.
The web‐based functional annotation tool, Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (http://david.abcc.ncifcrf.gov/tools.jsp), contains key components for disease, gene ontology, and pathway analyses.
Signaling pathway mapping of lung cancer‐related miRNAs
The signaling pathways and processes were explored using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Mapper (http://www.genome.jp/kegg/tool/map_pathway2.html), which is a collection of tools for KEGG mapping: KEGG pathway, BRITE, and MODULE mapping. The KEGG database consists of 16 main databases: systems information: KEGG PATHWAY, KEGG BRITE, KEGG MODULE, KEGG DISEASE, KEGG DRUG, and KEGG ENVIRON; genomic information: KEGG ORTHOLOGY, KEGG GENOME, KEGG GENES, KEGG SSDB, and KEGG; and chemical information: KEGG COMPOUND, KEGG GLYCAN, KEGG REACTION, KEGG RPAIR, KEGG RCLASS, and KEGG ENZYME.
Gene Expression Omnibus data analysis
The Gene Expression Omnibus (GEO) is a public functional genome data repository that supports MIAME‐compliant data submission. We used GEO2R as an interactive network tool, which allows users to compare two or more samples in a GEO sequence in order to identify genes expressed differently under different experimental conditions. The results are presented as a table of genes ordered by significance.
Results
Differential miRNA expression between smoking and non‐smoking lung cancer patients
Using GEO (GSE53519) database analysis, we compared the miRNA array data between NSCLC patients: smokers (10 samples), former smokers (10 samples), and non‐smokers (9 samples, control). There were 46 miRNAs, including 26 upregulated (P < 0.05, fold > 2) and 20 downregulated (P < 0.05, fold > 2) in the smoker compared to the non‐smoker group. Many of the miRNAs changed, including 6 upregulated and 29 downregulated miRNAs in the former smoker group compared to the non‐smoker group (Fig 1a,b). We selected the four common upregulated miRNAs (hsa‐mir‐133b, hsa‐mir‐634, hsa‐miR‐602, and hsa‐mir‐891a) and 16 downregulated miRNAs (hsa‐mir‐185, hsa‐mir‐548, hsa‐mir‐595, hsa‐mir‐613, hsa‐mir‐302f, hsa‐mir‐4288, hsa‐mir‐1294, hsa‐mir‐3172, hsa‐mir‐4295, hsa‐mir‐4297, hsa‐mir‐4307, hsa‐mir‐1282, hsa‐mir‐593, hsa‐mir‐1246, hsa‐mir‐1279, and hsa‐mir‐4308) from the Venny database (Fig 1c,d). Differential miRNAs were mainly downregulated in the former smoker group compared to the non‐smoker group. There were more downregulated miRNAs than upregulated miRNAs.
Figure 1.

(a,b) Differential and (c,d) commonly upregulated and downregulated microRNAs in the smoker versus non‐smoker groups and former smoker versus non‐smoker groups.
Differential miRNA regulated cell function
Based on the mirPath v.3 database, we analyzed the 4 upregulated and 16 downregulated miRNAs. Gene Ontology (GO), organelle, and ion binding classifications of the differential miRNAs showed that hsa‐mir‐133b and hsa‐mir‐602, and hsa‐mir‐891a and hsa‐mir‐634 had similar functions (Fig 2a). When the regulated messenger RNAs (mRNAs) of the down‐expressed miRNAs were classified (Fig 2b), we found three main categories: (i) hsa‐mir‐302f, hsa‐mir‐548, hsa‐mir‐4307, hsa‐mir‐4288, hsa‐mir‐1294, and hsa‐mir‐1246; (ii) hsa‐mir‐613, hsa‐mir‐4295, hsa‐mir‐4308, hsa‐mir‐4297, and hsa‐mir‐185‐3p; and (iii) hsa‐mir‐593‐3p, hsa‐mir‐1282, hsa‐mir‐595, and hsa‐mir‐1279. The functions of the regulated mRNAs by the down‐expressed miRNAs were mainly organelle, biosynthetic, ion binding, catabolic, and small molecular metabolic processes, and nucleic acid binding transcription factor activity.
Figure 2.
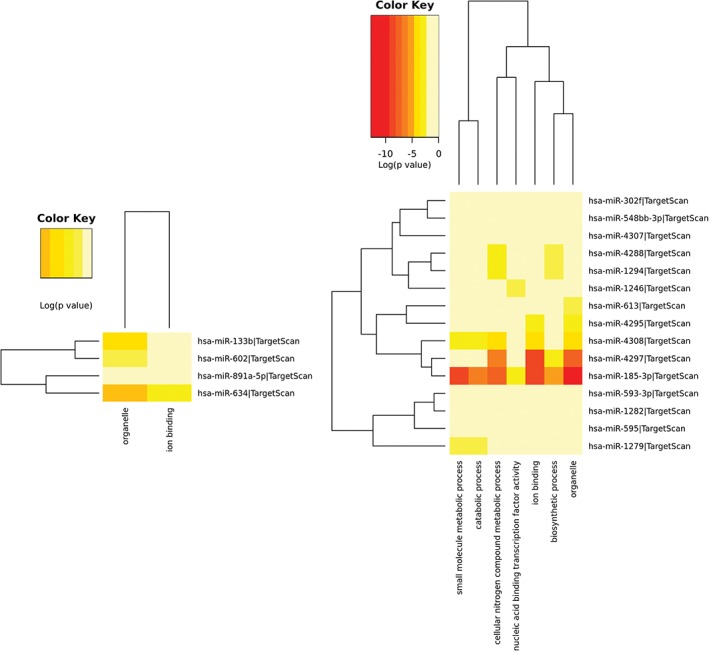
The function of (a) overexpressed and (b) down‐expressed microRNAs regulating messenger RNAs.
miRNAs affect the cell pathways by inhibiting messenger RNA expression
We used KEGG to analyze the target genes of the miRNA pathway. We found that the overexpressed miRNA‐related gene pathways were not associated with smoking induced drug resistance. The main pathways included Huntington's disease, adrenergic signaling in cardiomyocyte, endocrine, and other factor‐regulated calcium reabsorption. This indicated that down‐expressed miRNAs affect gene pathway communication, which causes a smoking induced response to therapy. The main pathways include nicotine addiction, the ERBB signaling pathway, endocrine and other factor‐regulated calcium reabsorption, and steroid hormone biosynthesis (Table 1). The signal pathway regulated by nicotine is strongly associated with smoking. The expression of many of the genes in these pathways was altered as a result of smoking, including CACNA1B, CHRNA4, GABRB2, SLC17A7, and GRIA1. These genes affect nicotine addiction. Each gene was regulated by one miRNA: hsa‐mir‐185‐3p interacted with CACNA1B, GRIA1, and CHRNA4; hsa‐mir‐1294 interacted with GABRB2; and hsa‐mir‐4308 interacted with SLC17A7. The down‐expression of these miRNAs increased the expression of these genes, making the pathway more active (Fig 3).
Table 1.
Overexpressed and down‐expressed miRNAs regulate target gene pathways
| KEGG pathway | P | Genes | miRNAs | KEGG pathway | P | Genes | miRNAs | ||
|---|---|---|---|---|---|---|---|---|---|
| Down | Steroid hormone biosynthesis | 3.22E‐09 | 11 | 3 | Up | Huntington's disease | 2.26E‐06 | 7 | 3 |
| Nicotine addiction | 0.005 | 9 | 6 | Adrenergic signaling in cardiomyocytes | 0.001 | 9 | 3 | ||
| Tyrosine metabolism | 0.011 | 5 | 1 | Parkinson's disease | 0.002 | 3 | 2 | ||
| Endocrine and other factor‐regulated calcium | 0.013 | 8 | 5 | Endocrine and other factor‐regulated calcium reabsorption | 0.02 | 3 | 2 | ||
| ErbB signaling pathway | 0.018 | 19 | 7 | Circadian entrainment | 0.03 | 3 | 3 |
miRNA, microRNA.
Figure 3.
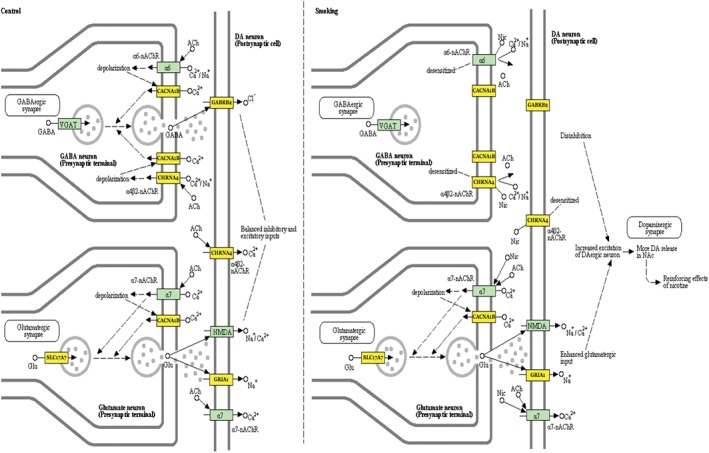
MicrRNAs regulate the smoking‐related nicotine addiction pathway.
Regulation of ERBB pathway activity by miRNAs through the ERBB signaling pathway
EGFR mutation in NSCLC is the main therapeutic target; however, smokers and former smokers are often drug resistant. We analyzed the miRNA data using TarBase version 7.0 and found that differential miRNAs primarily affect the targeted drug pathway by regulating gene expression in smoking patients. The DIANA‐TarBase was first released in 2006 with the aim of cataloguing experimentally validated miRNAs.19 Eighteen target genes were upregulated, while the downregulated miRNAs included GSK3B, PRKCA, ERBB2, SOS2, CAMK2G, CRK, RPS6KB2, PAK1, AKT2, PLCG1, PIK3CD, PIK3R3, MYC, PAK4, PAK6, ABL2, MAPK10, and EREG. Upregulation of these genes will induce ERBB2 pathway activity and block EGFR targeted drug activity. When we analyzed the mRNAs regulated by miRNAs, we found that hsa‐mir‐183‐3p affected the expression of other genes in this pathway, including PAK1, PAK4, PAK6, PIK3CD, PIK3R3, AKT2, ERBB2, PRKCA (1249), and CAMK2G(4297). Hsa‐mir‐4295 and hsa‐mir‐4288 have two targeted genes, hsa‐mir‐4295 regulates SOS2 and EREG, and hsa‐mir‐4288 regulates MAPK10 and GSK3B. ABL2, MYC, PLCG1, and CRK are regulated by hsa‐mir‐4297, hsa‐mir‐1294, hsa‐mir‐4308, and hsa‐mir‐1282, respectively. These results suggest that interaction of these seven miRNAs could regulate targeted drug resistance in smokers (Fig 4).
Figure 4.
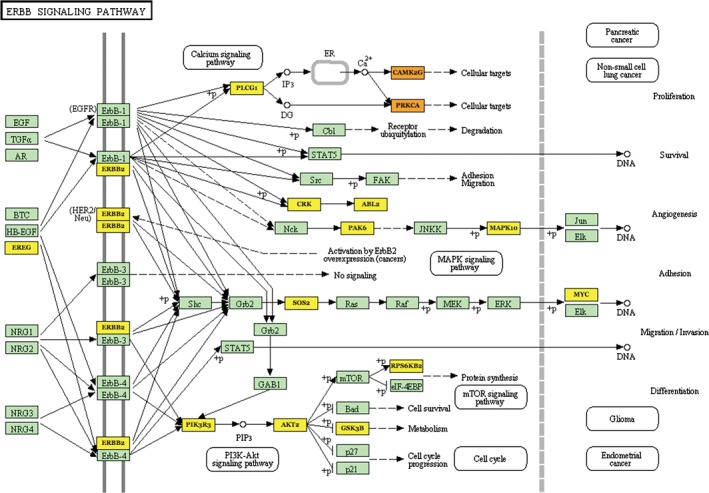
MicroRNAs regulate the smoking‐related ERBB2 signaling pathway.
miRNAs affect survival in smokers
In order to understand the effect of targeted genes in NSCLC patients, we analyzed the relationship between survival and gene expression by using the Kaplan–Meier Plotter database. We analyzed smoking‐related nicotine addiction pathway genes and found two aspects to the relationship between survival and gene expression. First, the higher the expression of genes such as CACNA1B, CHRNA4, and SLC17A7, the shorter the survival, suggesting that expression of these genes causes death. Second, the higher the expression of genes such as GRIA1 and GABRB2, the longer the survival, suggesting that expression of these genes promotes survival (Fig 5a). Thus, the genes in the ERBB signaling pathway have three different functional types: one group causes death (GSK3B, PRKCA, ERBB2, SOS2, CAMK2G, PAK1, AKT2, MYC, PAK4, PAK6, and ABL2); a second promotes survival (PLCG1, PIK3CD, and PIK3R3); and a third group has no relationship to survival (CRK, MAPK10, CRK, and EREG) (Fig 5b). These results show that the miRNAs that regulate the targeted genes are primarily in the first group, in which gene expression causes death. Down‐expression of these miRNAs also promotes drug resistance in NSCLC.
Figure 5.
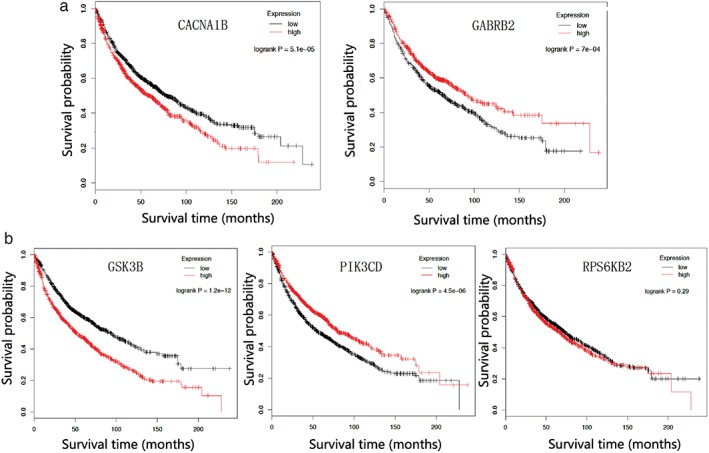
The relationship between microRNA targeted genes and survival. (a) Nicotine addiction and (b) ERBB signaling pathways. Expression ( ) low and (
) low and ( ) high.
) high.
Discussion
Lung cancer is a multistep process that transforms normal cells into malignant derivatives, leading to the conclusion that research into carcinogenesis from a systems perspective is unavoidable.20 Evidence of the association between cigarette smoking and cancers of the respiratory system (including the lung), digestive tract, and urinary system has accumulated over time.
Our results show that tobacco use induces the downregulation of many miRNAs, resulting in an increase in miRNA target gene expression levels. Gene pathway enrichment of miRNA target genes showed that the nicotine addiction and ERBB pathways were clearly altered. The largest proportion of target genes promotes death if their gene levels increase. The interaction network among hsa‐mir‐185‐3p, hsa‐mir‐4295, hsa‐mir‐4288, and hsa‐mir‐613 promotes lung cancer occurrence by affecting the ERBB pathway. At the same time, these miRNAs, including hsa‐mir‐185‐3p, hsa‐mir‐1294, and hsa‐mir‐4308, strengthen nicotine addiction by affecting their target genes.
Although we found that the common action of these miRNAs regulated NSCLC occurrence via the ERBB pathway in smokers, few articles have examined a relationship between these miRNAs and NSCLC. Studies have shown that many of the carcinogens produced by smoking increase aging, accompanied by molecular and genomic changes through genetic mutations or modifications.21 Oncogenic miRNAs suppress the expression of many tumor inhibitor molecules and promote accelerated aging with associated carcinogenesis; by contrast, suppression of these miRNAs decreases cell proliferation and strengthens cellular senescence. For example, miR‐126 has a crucial function as a cancer‐suppressive miRNA in different kinds of tissues, including the pancreas, liver, and lung.22 In lung carcinoma cells, miR‐126 expression is inhibited, leading to the upregulation of target genes encoding VEGF.23 Smoking has a significant effect on the expression of 133 miRNAs in the lung tissue of rats, and some of these miRNAs may be involved in carcinogenesis.24 In the normal human bronchial epithelial cells and lung cancer cells of smokers, smoking‐mediated miRNA changes in in vitro experimental model systems have confirmed the inhibition of miR‐48‐7b, which leads to the upregulation of ZEST 12 (SUZ12), BCE inhibitors,25 BMI1, WNT5A, MYC, and KRAS, thus increasing the proliferation, invasion, tumorigenicity, and metastasis potential of lung cancer cells.25 Among smokers, the expression of Lit‐7 family members is negatively correlated with the number of cigarettes smoked per day.26 The expression of carcinogenic miR‐21 is related to the number of daily cigarettes smoked in squamous cell carcinoma but not in adenocarcinoma.27 Interestingly, there was no significant difference in the history of smoking in this study, such as current and previous smoking status, the number of years of smoking, the age at beginning smoking, or the risk of passive smoking. Microarray analysis of RNA in bronchial epithelial cells showed significant differences in the expression of 28 miRNAs between smokers and non‐smokers.28 Interestingly, smokers have a higher risk of EGFR mutations (exons 18, 19, and 21) than non‐smokers.29 It has been suggested that the histological type of a tumor is closely related to the mechanism of smoking‐related carcinogens.
In conclusion, our results provide evidence to explain the mechanisms of lung cancer development in smokers.
Disclosure
No authors report any conflict of interest.
References
- 1. Filippi AR, Di Muzio J, Badellino S et al. Locally‐advanced non‐small cell lung cancer: Shall immunotherapy be a new chance? J Thorac Dis 2018; 10 (Suppl. 13): S1461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greillier L, Cortot AB, Viguier J et al. Perception of lung cancer risk: Impact of smoking status and nicotine dependence. Curr Oncol Rep 2018; 20 (Suppl. 1): 18. [DOI] [PubMed] [Google Scholar]
- 3. Weng MW, Lee HW, Park SH et al. Aldehydes are the predominant forces inducing DNA damage and inhibiting DNA repair in tobacco smoke carcinogenesis. Proc Natl Acad Sci U S A 2018; 115 (27): E6152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li AR, Chitale D, Riely GJ et al. EGFR mutations in lung adenocarcinomas: Clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn 2008; 10 (3): 242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Merrick DT, Kittelson J, Winterhalder R et al. Analysis of c‐ERBB1/epidermal growth factor receptor and c‐ERBB2/HER‐2 expression in bronchial dysplasia: Evaluation of potential targets for chemoprevention of lung cancer. Clin Cancer Res 2006; 12 (7 Pt 1): 2281–8. [DOI] [PubMed] [Google Scholar]
- 6. Sakuma Y, Matsukuma S, Yoshihara M et al. Epidermal growth factor receptor gene mutations in atypical adenomatous hyperplasias of the lung. Mod Pathol 2007; 20 (9): 967–73. [DOI] [PubMed] [Google Scholar]
- 7. Song H, Sun B, Liao Y et al. GPRC5A deficiency leads to dysregulated MDM2 via activated EGFR signaling for lung tumor development. Int J Cancer 2019; 144 (4): 777–87. [DOI] [PubMed] [Google Scholar]
- 8. Li Z, Guo H, Lu Y, Hu J, Luo H, Gu W. Chemotherapy with or without pemetrexed as second‐line regimens for advanced non‐small‐cell lung cancer patients who have progressed after first‐line EGFR TKIs: A systematic review and meta‐analysis. Onco Targets Ther 2018; 11: 3697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Díaz‐Serrano A, Gella P, Jiménez E, Zugazagoitia J, Paz‐Ares Rodríguez L. Targeting EGFR in lung cancer: Current standards and developments. Drugs 2018; 78: 893–911. [DOI] [PubMed] [Google Scholar]
- 10. Mar N, Vredenburgh JJ, Wasser JS. Targeting HER2 in the treatment of non‐small cell lung cancer. Lung Cancer 2015; 87 (3): 220–5. [DOI] [PubMed] [Google Scholar]
- 11. Yasugi M, Takigawa N, Ochi N et al. Everolimus prolonged survival in transgenic mice with EGFR‐driven lung tumors. Exp Cell Res 2014; 326 (2): 201–9. [DOI] [PubMed] [Google Scholar]
- 12. Ambros V. The functions of animal microRNAs. Nature 2004; 431 (7006): 350–5. [DOI] [PubMed] [Google Scholar]
- 13. Wang Q, Li DC, Li ZF et al. Upregulation of miR‐27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene 2011; 30 (36): 3875–86. [DOI] [PubMed] [Google Scholar]
- 14. Sato T, Shiba‐Ishii A, Kim Y et al. miR‐3941: A novel microRNA that controls IGBP1 expression and is associated with malignant progression of lung adenocarcinoma. Cancer Sci 2017; 108 (3): 536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li B, Ding CM, Li YX, Peng JC, Geng N, Qin WW. MicroRNA‐145 inhibits migration and induces apoptosis in human non‐small cell lung cancer cells through regulation of the EGFR/PI3K/AKT signaling pathway. Oncol Rep 2018; 40 (5): 2944–54. [DOI] [PubMed] [Google Scholar]
- 16. Guo N, Zhao Y, Zhang W, Li S, Li S, Yu J. MicroRNA‐133a downregulated EGFR expression in human non‐small cell lung cancer cells via AKT/ERK signaling. Oncol Lett 2018; 16 (5): 6045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qi Z, Zhang B, Zhang J et al. MicroRNA‐30b inhibits non‐small cell lung cancer cell growth by targeting the epidermal growth factor receptor. Neoplasma 2018; 65 (2): 192–200. [DOI] [PubMed] [Google Scholar]
- 18. Wang Y, Zhao M, Liu J, Sun Z, Ni J, Liu H. miRNA‐125b regulates apoptosis of human non‐small cell lung cancer via the PI3K/Akt/GSK3β signaling pathway. Oncol Rep 2017; 38 (3): 1715–23. [DOI] [PubMed] [Google Scholar]
- 19. Vlachos IS, Paraskevopoulou MD, Karagkouni D et al. DIANA‐TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 2015; 43 (Database issue): D153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang YC, Chen BS. A network‐based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Med Genomics 2011; 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagasaki T1, Matsumoto H. Influences of smoking and aging on allergic airway inflammation in asthma. Allergol Int 2013; 62: 171–9. [DOI] [PubMed] [Google Scholar]
- 22. Ebrahimi F, Gopalan V, Smith RA, Lam AKY. miR‐126 in human cancers: Clinical roles and current perspectives. Exp Mol Pathol 2014; 96 (1): 98–107. [DOI] [PubMed] [Google Scholar]
- 23. Kong R, Ma Y, Feng J et al. The crucial role of miR‐126 on suppressing progression of esophageal cancer by targeting VEGF‐A. Cell Mol Biol Lett 2016; 21: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Izzotti A, Larghero P, Longobardi M et al. Dose‐responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat Res 2011; 717 (1–2): 9–16. [DOI] [PubMed] [Google Scholar]
- 25. Xi S, Xu H, Shan J et al. Cigarette smoke mediates epigenetic repression of miR‐487b during pulmonary carcinogenesis. J Clin Invest 2013; 123 (3): 1241–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Exley C, Begum A, Woolley MP, Bloor RN. Aluminum in tobacco and cannabis and smoking‐related disease. Am J Med 2006; 119 (3): 276.e9–11. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Pan T, Zhong X, Cheng C. Nicotine upregulates microRNA‐21 and promotes TGF‐β‐dependent epithelial‐mesenchymal transition of esophageal cancer cells. Tumour Biol 2014; 35 (7): 7063–72. [DOI] [PubMed] [Google Scholar]
- 28. Chari R, Lonergan KM, Ng RT, MacAulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics 2007; 8: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosell R, Taron M, Reguart N, Isla D, Moran T. Epidermal growth factor receptor activation: How exon 19 and 21 mutations changed our understanding of the pathway. Clin Cancer Res 2006; 12: 7222–31. [DOI] [PubMed] [Google Scholar]


