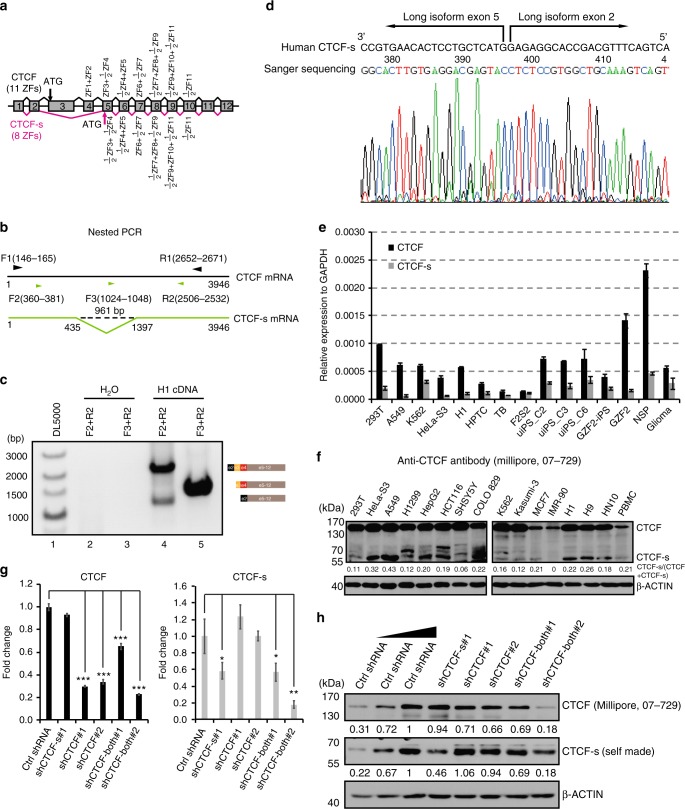
Fig. 1.
Identification of an alternatively spliced CTCF-s isoform in the human genome. a Schematic representation of exons of the human CTCF gene. Transcripts including alternative exons 3 and 4 encoded the widely expressed canonical form of CTCF (top), and the CTCF-s isoform excluded exons 3 and 4. The canonical form of CTCF contained 11 ZFs, while CTCF-s effectively lacked the N terminal and 3 ZFs. b Strategy for detecting the CTCF short isoform. Black line represents the full length of CTCF mRNA (1−3946), and green line represents CTCF-s mRNA (1−435 joined with 1397−3946). Primer information was indicated based on the canonical long isoform. Convergent black arrows showed the positions of the first nested primers; green arrows indicated the position of the second nested PCR primers, with F2 and R2 for amplifying both CTCF and CTCF-s, F3 and R2 for amplifying CTCF only. c Nested PCR was used to validate the existence of the short isoform with the primers from (b). The constitution of each PCR product is presented on the right. d Chromatogram from Sanger sequencing of the lower band of panel (c), showing the junction at exons 2 and 5. e TaqMan RT-qPCR analysis of the relative expression levels of CTCF and CTCF-s in various human cell lines. Data were shown as mean ± s.d., n = 3 technological repeats. f Western blot of CTCF and CTCF-s in different human cell lines with anti-CTCF antibody (Millipore, 07-729). The locations of the full-length CTCF and CTCF-s were indicated. g RT-qPCR analysis of two different CTCF isoforms after specific shRNA knockdown. Data were shown as mean ± s.d. from n = 3 independent technological repeats with the indicated significance by using a two-tailed Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001. h Western blot analysis of two different CTCF isoforms after specific shRNA knockdown. Source data are provided as a Source Data file