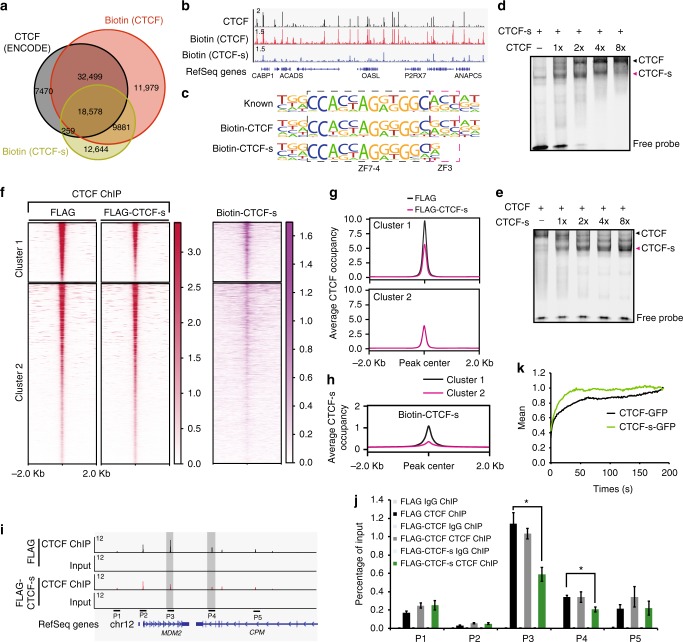
Fig. 2.
CTCF-s has its specific genomic binding but also competes with CTCF in binding to their common targets. a Venn diagram showing the overlap of biotin-enriched CTCF or CTCF-s peaks with a normal CTCF ChIP-seq from ENCODE (GSE33213). b Genomic views of ChIP-seq intensities of CTCF (ENCODE), biotin ChIP-seq for biotin-CTCF and biotin-CTCF-s in HeLa-S3 cells at chromosome 12:121,055,606−121,842,881. c Consensus DNA binding motif generated from de novo motif discovery. The motif inside the black dotted box is bound by ZF4-7 of CTCF and the motif inside the magenta dotted box is bound by ZF3 of CTCF. d Electrophoresis mobility shift analysis (EMSA) showing DNA binding ability of CTCF and CTCF-s as the concentrations of CTCF increase. e EMSA showing DNA binding ability of CTCF and CTCF-s with increasing amounts of CTCF-s. f Heatmap showing the comparison of CTCF binding occupancies between FLAG control and FLAG-CTCF-s overexpression. Cluster 1 shows the significantly reduced occupancy of CTCF after CTCF-s overexpression; Cluster 2 shows no difference in the levels of CTCF genomic occupancy with or without CTCF-s overexpression. Heatmap on the right showing the biotin ChIP-seq signal for CTCF-s. g Tag density pileup for all CTCF peaks from clusters 1 and 2, respectively. h Tag density pileup for biotin-CTCF-s peaks from clusters 1 and 2, respectively. i Genomic view showing ChIP primers P1−P5 at MDM2-CPM loci in chromosome 12. Peaks marked by gray box represent two decreased CTCF peaks upon CTCF-s gain-of-function. j Verification of CTCF ChIP-seq data in chromosome 12 using ChIP-qPCR. IgG was used as a negative control. The locations of the primer pairs P1−P5 are indicated in panel (i). The data were represented as mean ± s.d. (n = 3 technological repeats) with the indicated significance by using a two-tailed Student’s t test (*p < 0.05). The entire experiment was performed three times. k FRAP analysis of GFP-CTCF and GFP-CTCF-s after irreversible photobleaching. Source data are provided as a Source Data file