Fig. 8.
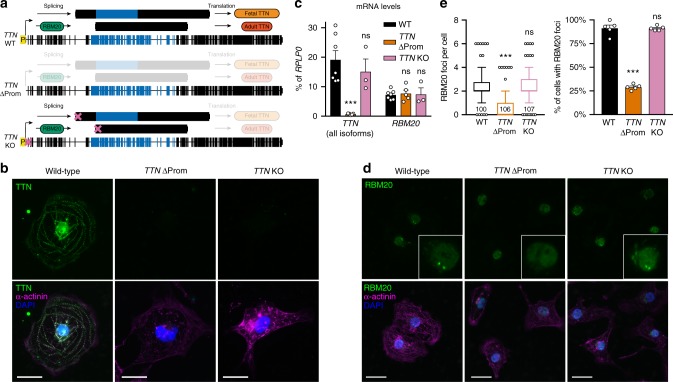
TTN transcription is required for nucleation of RBM20 into foci. a Schematic of the hESC gene editing strategies used to test the role of transcription at the TTN locus. ∆Prom: promoter (P) deletion; KO: functional knockout by frameshift mutation (indicated by the X). Aspects of TTN biogenesis predicted to be impaired by each modification are indicated by increased transparency (loss of transcription following promoter deletion; loss of translation following knockout). Alternatively spliced exons regulated by RBM20 are shown in blue. b Representative immunofluorescence results in CM derived from the indicated hESC lines (nuclei counterstained with DAPI); scale bars: 10 µm. c RT-qPCR in CM derived from the indicated hESC lines. Expression is relative to the housekeeping gene RPLP0 and mean ± s.e.m. is shown. n = 3, 5, or 7 independent differentiations for TTN KO, TTN ΔProm, and WT, respectively. d As in panel (b); insets show magnified views. e Quantification of RBM20 foci in CM derived from the indicated hESC lines. On the left, box and whiskers plots showing median, 25th and 75th percentile, and the 10–90 percentile range plus outliers; the number of cells is indicated (non-diploid CM were included in the analysis). On the right mean ± s.e.m.; n = 5 field of views. All p-values in this figure are calculated vs WT by one-way ANOVA followed by Holm-Sidak’s multiple comparisons (for (c) and the right graph in (e)) or by Kruskal–Wallis test followed by Dunn’s multiple comparisons (for the left graph in (e)); ns ≥ 0.05; *** < 0.01. Source data are provided as a Source Data file