Fig. 9.
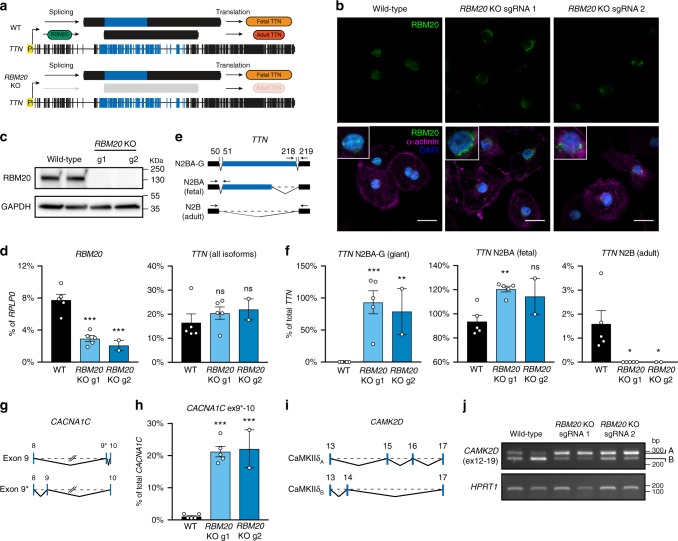
Knockout of RBM20 dysregulates alternative splicing in hESC-derived cardiomyocytes. a Predicted effects of the knockout of RBM20 on TTN biogenesis (loss of RBM20-dependent alternative exon exclusion). Refer to the legend of Fig. 8a for the abbreviations. b Representative immunofluorescence results in CM derived from wild-type or RBM20 knockout hESCs generated with two independent CRISPR/Cas9 single guide RNAs (sgRNAs). Nuclei are counterstained with DAPI, and insets show magnified views; scale bars: 10 µm. c Western blot validation of RBM20 knockout (uncropped images shown in Supplementary Fig. 11). g: sgRNA. d RT-qPCR in CM derived from the indicated hESC lines. Expression is relative to the housekeeping gene RPLP0 and mean ± s.e.m. is shown. n = 5 (WT and RBM20 KO g1) or 2 (RBM20 KO g2) independent differentiations. e Expected major TTN isoforms in CM. Exons predicted to be excluded by RBM20 are in blue, and arrows indicate the location of RT-qPCR primers. f As in panel (d), but expression of the indicated TTN isoform is relative to the total amount of TTN. g Expected CACNA1C isoforms due to RBM20-dependent alternative exon selection. RBM20 is predicted to favor inclusion of exon 9 over exon 9*. h As in panel (d), but expression of the CACNA1C exon 9* isoform is relative to the total amount of CACNA1C. i Expected CAMK2D isoforms due to RBM20-dependent alternative exon selection. RBM20 is predicted to favor inclusion of exon 14 over exons 15–16. j Semiquantiative RT-PCR in CM for the indicated CAMK2D isoforms and the housekeeping gene HPRT1; n = 2 independent differentiations. All p-values in this figure are vs WT and calculated by one-way ANOVA followed by Holm-Sidak’s multiple comparisons (ns ≥ 0.05; * < 0.05; ** < 0.01; *** < 0.001). Source data are provided as a Source Data file