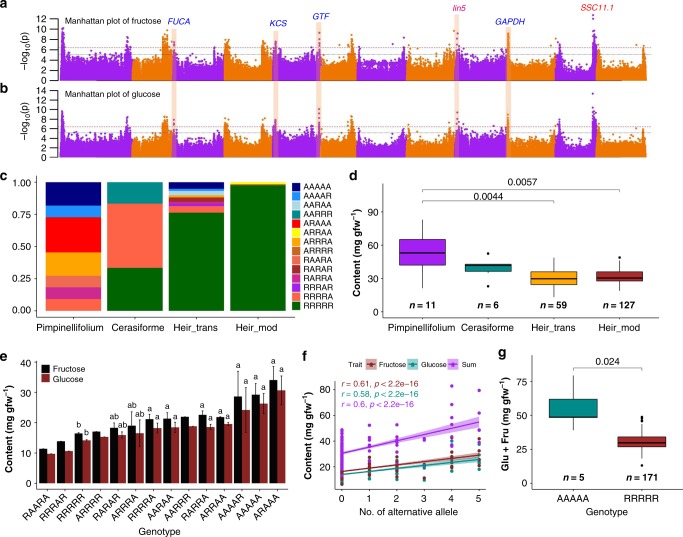
Fig. 2.
Combinations of fructose and glucose alleles for the improvement of tomato sugar content. Only alleles that were significantly associated both with fructose and glucose were analyzed. a, b Manhattan plot for meta-analysis of genome-wide association analysis of fructose (a) and glucose (b) content. Candidates and previously identified genes were labeled in blue and red, respectively. FUCA, alpha-L-fucosidase 1; KCS, fatty acid elongase 3-ketoacyl-CoA synthase; GTF, glucosyltransferase; GADPH, glyceraldehyde-3-phosphate dehydrogenase. c Allele distribution of fructose/glucose content at positions: chr3:1,506,106, chr5:3,403,706, chr5:63,485,334, chr9:3,477,979, and chr10:422,707 that were both significantly associated with fructose and glucose in S. lycopersicum var cerasiforme (cerasiforme), heirloom + transitional (heir_trans), heir + modern (heir_mod), and the closest wild species S. pimpinellifolium (pimpinellifolium) tomato accessions (see detailed information about groups in online methods). d Comparison of sugar content (fructose + glucose) between different tomato types in cerasiforme, heir_trans, heir_mod, and pimpinellifolium tomato accessions. e Mean (±SE) content of fructose (black) and glucose (brown) at different allele combinations in cerasiforme, heir_trans, heir_mod, and pimpinellifolium tomato accessions. Significant t-test P values are also provided. f Correlation between the number of alternative alleles and sugar content. Fructose, glucose, and the sum of fructose + glucose were colored in brown4, cyan4, and purple. g Comparison of sugar content (fructose + glucose) between all alternative and reference allele combinations at position chr3: 1,506,106, chr5: 3,403,706, chr5: 63,485,334, chr9: 3,477,979, and chr10: 422,707. Center line and limits of box were the mean and interquartile ranges. Error bars represent the maximum and minimum values. Whiskers indicate variability outside the upper and lower quartiles. Significant t-test P values are also provided. Source data of Fig. 2c–g are provided in a Source Data file