Abstract
Our previous studies showed that in a mouse model in which PI3K-AKT activation was increased (YF mice), osteoclast numbers and levels of SDF-1, a chemokine, were augmented. The purpose of this study was to delineate the role of PI3K activation in regulating SDF-1 production and examine whether SDF-1 can stimulate differentiation and/or migration of osteoclast precursors. Using flow cytometric analysis, we demonstrated that compared to wild type mice, bone marrow of YF mice had increased numbers of CXCL12 abundant reticular (CAR) cells, that are a major cell type responsible for producing SDF-1. At the molecular level, transcription factor specificity protein 1 (Sp1) induced an increased transcription of SDF-1 that was dependent on PI3K/AKT activation. YF mice also contained an increased number of osteoclast precursors, in which expression of CXCR4, a major receptor for SDF-1, was increased. SDF-1 did not induce differentiation of osteoclast precursors into mature osteoclasts; compared to cells derived from WT mice, cells obtained from YF mice were more responsive to SDF-1. In conclusion, we demonstrate that PI3K activation resulted in increased SDF-1, increased the number of osteoclast precursors, and enhanced osteoclast precursor migration in response to SDF-1.
Keywords: Osteoclast precursors, CAR cells, SDF-1, PI3K, Migration
Highlights
-
•
PI3K activation regulates the number of CAR cells in mouse bone marrow.
-
•
PI3K activation regulates SDF-1/CXCL12 production by CAR cells in bone marrow.
-
•
PI3K/AKT activation mediates transcription of SDF-1 by regulating transcription factor Sp1.
-
•
SDF-1 enhances migration of osteoclast precursors via CXCR4.
1. Introduction
Stromal cell Derived Factor-1 (SDF-1) also known as CXC Ligand 12, (CXCL12) belongs to C-X-C family of chemokines, with the seven transmembrane G-protein coupled receptor, CXCR4 as its major receptor (Nagasawa, 2014; Nagasawa et al., 1996a). SDF-1 plays a critical role in the initiation and maintenance of bone marrow hematopoietic niche in mice (Greenbaum et al., 2013; Omatsu et al., 2010; Sugiyama et al., 2006). Mice deficient in SDF-1 or CXCR4 die in utero and have very few hematopoietic precursors and their progenitors in the bone marrow (Ma et al., 1999; Nagasawa et al., 1996b). SDF-1 is critical for cell migration. Hematopoietic stem cells, endothelial progenitor cells and lymphocytes depend on SDF-1 for their homing ability (Wang and Knaut, 2014). The SDF-1/CXCR4 pathway is implicated in osteoclast precursor migration, osteoclast differentiation and bone resorption (Grassi et al., 2004; Gronthos and Zannettino, 2007; Wright et al., 2005; Zhang et al., 2008).
Phosphatidylinositol‑3 Kinase (PI3K), a prominent lipid kinase, phosphorylates the 3′‑hydroxyl group on the inositol ring of phosphoinositdylinositol. While class II and III PI3Ks are constitutively activated, Class I PI3K are activated in response to extracellular stimuli. Class I PI3K is a heterodimer of p110 catalytic and p85 regulatory subunits. The p110 catalytic subunit binds to the p85 regulatory subunit with high affinity and this interaction prevents inadvertent activation of PI3K (Cantley, 2002; Engelman et al., 2006). Upon engagement of the SH2 domain of the p85 subunit through binding to phosphorylated tyrosines of receptors or signaling proteins, the inhibitory effect of p85 on p110 is released, resulting in increased lipid kinase activity (Foukas and Okkenhaug, 2003).
Cbl (Casitas B-lineage Lymphoma) protein is an E3 ubiquitin ligase and an adaptor, which regulates bone remodeling through its interaction PI3K (Adapala et al., 2010a; Brennan et al., 2011). Cbl directly binds to the p85 regulatory subunit of PI3K, via the tyrosine 737 in the YEAM motif, which is phosphorylated by tyrosine kinase c-Src (Miyazaki et al., 2004). A substitution of tyrosine to phenylalanine in the YEAM motif prevents phosphorylation of Cbl at Y737 position and abrogates Cbl-p85 interaction (Feshchenko et al., 1998, Feshchenko et al., 1999).
Our previous studies have shown that knock-in mice harboring the Y737F mutation (YF mice) have increased PI3K-AKT activation (Adapala et al., 2010a, Adapala et al., 2010b, Adapala et al., 2014a; Scanlon et al., 2017). YF mice increased bone volume due to a combination of increased non-resorptive osteoclasts and increased bone formation by osteoblasts (Adapala et al., 2010a; Brennan et al., 2011). SDF-1 has been shown to promote the proliferation of osteoblast precursors (Kortesidis et al., 2005; Zhu et al., 2011). We previously reported that proliferation of osteoblast precursors and levels of SDF-1 in the bone marrow were significantly increased in YF mice (Brennan et al., 2011).
Given our previous observations, we hypothesized that increased PI3K activation augmented SDF-1 production, stimulated SDF-1-mediated osteoclast precursor migration and increased the number of osteoclast precursors in the bone marrow. Using YF mice, we examined whether PI3K activity regulates SDF-1 production by CAR cells, the mechanism of increased SDF-1 production and the effect of SDF-1 on osteoclast precursors differentiation and migration.
2. Materials and methods
2.1. Mice
Generation of CblY737F/Y737F (YF) mice with a point mutation on the p85 regulatory subunit-binding site of Cbl were previously described (Molero et al., 2006). Mice were genotyped for WT and YF alleles as previously reported (Adapala et al., 2010a). All mice used in the study were in a mixed C57BL/6JX129SvJ background. For all experiments 6–8 week old male mice were used. All animal procedures were conducted according to protocols approved by the University of Connecticut Health Center Institution Animal Care and Use Committee.
2.2. Reagents
Minimum essential medium-α modification (α-MEM) and fetal bovine serum (FBS) were purchased from Sigma (St. Louis, MO), Penicillin/ Streptomycin from CellGro (Mediatech, Manassas, VA), RANK ligand (RANKL) and MCSF were purchased from R&D Systems (Minneapolis, MN). Leukocyte acid phosphatase kit for tartrate-resistant acid phosphatase (TRAP) staining of osteoclasts, Calcein, LY294002, AMD3100, AKT inhibitor and Mithramycin were purchased from Sigma (St. Louis, MO). Transwell chambers for migration assay were purchased from Corning (Tewksbury, MA). For western blotting analysis, anti-phospho-AKT (Thr308), anti-AKT, anti-phospho GSKα/β Ser21/9, anti-GSK-beta, anti-Actin, anti-Tubulin antibodies were purchased from Cell Signaling Technology (Danvers, MA) and anti-Sp1 antibody, IgG secondary antibody from Millipore (Temecula, CA). HBSS, HEPES and fetal calf serum were purchased from Gibco, Invitrogen (Carlsbad, CA). Rhodamine phalloidin actin stain was purchased from Invitrogen (Carlsbad, CA). Immunofluorescence images were captured using a Leica fluorescent microscope (Buffalo Grove, IL).
2.3. Antibodies
Monoclonal antibodies were used for phenotyping the cell surface markers for identification of specific populations, sorting of CAR cells, non-hematopoietic, and non-endothelial bone marrow reticular cells. All antibodies, conjugated to fluorochrome or biotinylated, were purchased from e-Biosciences (San Diego, CA), Biolegend (San Diego, CA), Pharmingen/BD Bioscience (San Diego, CA), or R&D Systems (Minneapolis, MN) unless specified otherwise. For CAR cells: anti-CD45/Ly-5 (clone 30-F11), anti-Ter119/Ly-76 (TER-119), anti CD31/PECAM-1 (clone 390), anti- Sca-1/Ly-6A/E (clone D7), anti-CD140b/PDGF receptor b (clone APB-5) and anti-CD106/VCAM-1 (clone 429). For SDF-1 production and CXCR4 expression: anti-mouse IgG1 FITC isotype, anti-human/mouse CXCL12/SDF-1 (aa 20–89), Anti-CD184/CXCR4 (clone 2B11). For osteoclast precursors: anti-c-Fms/CD115 (clone AFS98) was used. For magnetic sorting of CD45- Ter119- CD31- Sca-1- population anti-PE micro beads (Miltenyi, Auburn, CA) were used and desired population was isolated using manufacturer's protocol. Propidium Iodide used for the exclusion of dead cells was purchased from Sigma (St. Louis, MO).
2.4. Flow cytometric analysis
Bone marrow was flushed from femora and tibiae using a 25-gauge needle. RBCs were lysed using ACK (0.15 M ammonium chloride, 1 M potassium bicarbonate, 0.1 M EDTA) lysis buffer. Cells were washed and filtered through 100 μm Nytex mesh to remove cell clumps. The labeling for flow cytometric analysis or cell sorting was performed by standard staining procedures. Antibody staining was performed with optimal dilutions of directly conjugated or biotinylated monoclonal antibodies and secondary antibodies in sequential steps. Cell staining was performed on ice using staining medium containing 1 HBSS, 0.01 M HEPES (pH 7.4) and 2% fetal calf serum. Cells were treated with propidium iodide (10 μg/ml) to identify and exclude dead cells. Flow cytometric Flow analysis and sorting was performed in a BD-FACS Aria (BD Biosciences, San Jose, CA, USA) equipped with five lasers and 18 fluorescence detectors. All the data was analyzed using FlowJo software from Tree Star Inc. (Ashland, OR, USA).
2.5. Cell culture
For generation of osteoclast (OC), bone marrow was isolated from long bones of 4–6-week-old mice. Following overnight incubation on tissue culture plastic, non-adherent cells were plated at 2.5 × 105/cm2 in α-MEM, 10% FBS, penicillin/streptomycin containing 20 ng/ml M-CSF. Subsequently, cells were treated with M-CSF (20 ng/ml) and RANKL (50 ng/ml) in the presence or absence of SDF-1 (100 ng/ml) or AMD3100 (10 ng/ml) for an additional 5–6 days. Following TRAP staining, osteoclasts were identified as TRAP stained cells with >3 nuclei per cell.
2.6. RT PCR analysis
In some experiments non-hematopoietic, non-endothelial cells were obtained from bone marrow by magnetic sorting of CD45− Ter119− CD31− Sca1− cells using anti-PE micro-beads (Miltenyi Inc. Auburn, CA) following manufacturer's protocol. Cells were cultured in Minimum essential medium-α modification (α-MEM) containing 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin and harvested for RNA isolation as previously reported (Wang and Knaut, 2014). In some experiments, cells were treated with Mithramycin, for 3 h and RNA isolation was performed using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. A Super Script II reverse transcriptase kit (Invitrogen) was used according to manufacturer's instructions. Oligo (dT) and dNTPs were purchased from Promega (Madison, WI). Quantitative real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 7500 real-time PCR system. Gene expression levels were normalized to GAPDH and calculated using the DDCt method. Primers were designed using mRNA sequences obtained from Mouse Genomics Informatics and the Primer3 program. Following primers were used: SDF-1, AGTAGTGGCTCCCCAGGTTT (sense) and GAGACAGTCTTGCGGACACA(antisense); GAPDH, AACTTTGGCATTGTGGAAGG (sense) and ACACATTGGGGGTAGGAACA (antisense).
2.7. Migration assay
Total bone marrow cells were plated on petri dishes in the presence of MCSF (20 ng/ml) and cultured for 3 days to generate bone marrow macrophages (BMMS). BMMs were cultured with MCSF (20 ng/ml) and 50 ng/ml of RANKL to generate osteoclast precursors (OCPs). OCPs detached were detached with Accutase (Sigma, St. Louis, MO) and placed in the upper chamber of transwell plate with or without AMD3100 (10 μg/ml). SDF-1 was added to the lower chamber at 100 ng/ml. Following migration for 3 h, cells were in the lower chamber were incubated with calcein and the number of cells migrated to lower chamber as a percentage of total number of cells added to upper chamber was determined by fluorometric reading using BioTek Synergy HT (Winooski, Vermont).
2.8. Evaluation of biochemical activities
Cells were cultured in medium devoid of serum and growth factors for 60 min and then stimulated with SDF-1 (100 ng/ml). In some experiments, prior to the addition of the SDF-1, cells were treated with AMD3100 or LY294002 for 30 min. Total cell lysate was subjected to SDS-PAGE electrophoresis and transferred to Whatman PROTRAN nitrocellulose membrane (Whatman GmbH, Dassel, Germany). Nuclear/cytoplasmic lysates were prepared using Active Motif nuclear extract kit (Active Motif, Carlsbad, CA). Blots were probed for phosphorylation of AKT or GSK stripped and reprobed for total AKT or GSK-beta using specific antibodies. Blots were probed using Infrared conjugated LICOR secondary antibodies. The amounts of total proteins in individual bands were quantified by using Odyssey Infrared Imaging Systems software 2.1 (LICOR Biosciences, Lincoln, Nebraska).
2.9. Statistical analysis
Based on our previous studies we calculated a sample size of at least 10 mice per group (effect size =1.5, α = 0.05, β = 0.1, power = 90%). Experiments conducted in this study were repeated at least three times. Statistical significance between 2 groups were determined using Student's t-test, whereas for >2 groups, Analysis of variance (ANOVA) with Tukey's multiple comparison tests was performed using Graph Pad Prism (La Jolla, CA). Data are presented as means ± SEM. p-values <0.05 vs. control were considered statistically significant.
3. Results
3.1. In YF mice number of CAR cells and SDF-1 expression in bone marrow is increased
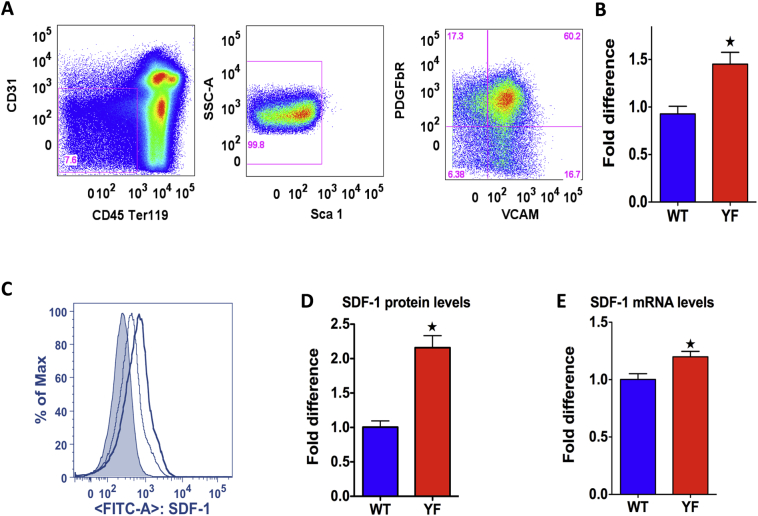
CXCL12 abundant reticular (CAR) cells are a major source of SDF-1 production. (Greenbaum et al., 2013; Omatsu et al., 2010; Sugiyama et al., 2006). CAR cells express vascular cell adhesion molecule-1 VCAM-1/CD103, CD44+ and platelet-derived growth factor β and are identified as CD45−CD31−SCa−1- CD103+ and CD140β+ (Sugiyama et al., 2006; Nagasawa et al., 2011). They do not express endothelial marker PECAM-1/CD31 or smooth muscle cell marker smooth muscle α-actin (SMA), which indicate that they are different from endothelial and smooth muscle cells (Tokoyoda et al., 2004). Here we phenotypically characterized as CD45−Ter119−CD31−Sca1−VCAM-1+PDGFbR+ (Fig. 1A). We performed flow analysis to identify CAR cells from bone marrow of WT and YF mice. In YF mice there was a 50% increase in the number of CAR cells in bone marrow (Fig. 1B) and there was 120% increase in the intracellular levels of SDF-1 protein as determined by mean fluorescence intensity (Fig. 1C and D). Increased SDF-1 level was also confirmed by determining SDF-1 mRNA expression in sorted CAR cells. Compared to the WT, there was a 20% increase in mRNA levels of SDF-1 in YF cells (Fig. 1E). In contrast the level of SDF-1 was not altered between the two genotypes in endothelial cells (CD31+ cells), which are also a major source of SDF-1 (Suppl. Fig. 1A and B).
Fig. 1.
Number of CXCL12 abundant reticular cells and SDF-1 production in YF bone marrow. Flow analysis of CAR cells and SDF-1 production. A. Total bone marrow cells were gated for a population negative for CD45, Ter119, CD31, and Sca1 and positive for VCAM and Pdgfr, the phenotype of CAR cells. B. Number of CAR cells in bone marrow of 8 week-old WT and YF male mice. Percentage of CAR cells in bone marrow was calculated as the number in YF bone marrow relative to WT bone marrow. Data is pooled from five independent experiments (WT, n = 25; YF n = 27). C. Intracellular level of SDF-1 was determined by the mean fluorescence intensity (factor of mean SDF-1 staining and SDF-1 positive cell count). Histogram in grey shade represents mouse IgG FITC isotype control. Bold black line represents SDF-1 FITC stain in YF cells and thin grey line represents in WT cells. D. SDF-1 intracellular level is shown as amount of protein in CAR cells in YF bone marrow relative to CAR cells in WT bone marrow (n = 9 mice/genotype). E. qRT-PCR analysis of SDF-1 mRNA levels in FACS sorted CAR cells from bone marrow (n = 9 mice/genotype). *P value of <0.05 vs. WT.
3.2. PI3K/AKT/Sp1 axis mediate SDF-1 transcription
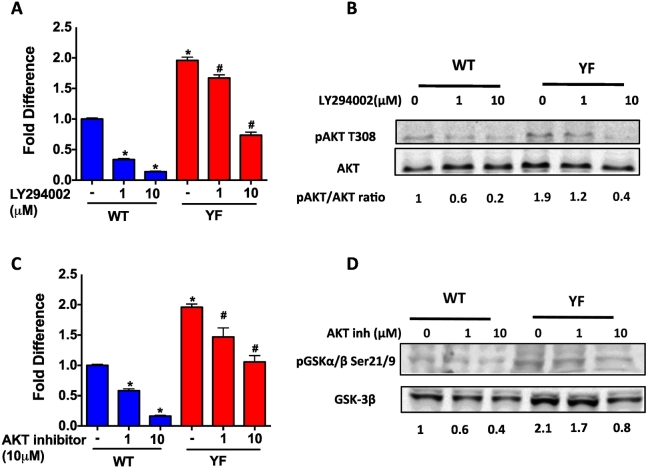
We performed a series of experiments to understand whether the increased SDF-1 expression is a result of increased PI3K activity. Sorted bone marrow cells were treated with LY294002 to inhibit PI3K activity. Our results showed that the basal level of SDF-1 gene expression was 2-fold higher in YF cells compared to WT cells (Fig. 2A). Inhibition of PI3K activity resulted in a decrease in SDF-1 gene expression in both WT and YF cells. However, inhibition of SDF expression in YF cells was observed only at higher dose of the inhibitor. The requirement of higher concentration of LY294002 to inhibit of SDF-1 gene expression corresponded to our finding that a higher dose of inhibitor was needed to attenuate pAKT levels in YF cells. Increased PI3K-AKT activity in YF cells compared to WT cells was confirmed by increased phosphorylation AKT (Fig. 2B). Similar results to the effects of LY294002 were obtained with an AKT inhibitor (Fig. 2C) and, the inhibition of AKT activity was confirmed by decreased phosphorylation of GSK (Fig. 2D), which is a substrate of AKT.
Fig. 2.
Increased PI3K activity and increased transcription of SDF-1 in the absence of Cbl-PI3K interaction. A. qRT-PCR analysis of SDF-1 mRNA levels in magnetically sorted CD45 Ter119 CD31 Sca1 negative population of bone marrow cells, cultured and treated with PI3K inhibitor LY294002 at indicated concentrations for 3 h. SDF-1 mRNA levels were normalized to GAPDH, and values shown are relative to the SDF-1 mRNA levels in untreated WT cells. B. Western blotting analysis of AKT phosphorylation, indicative of PI3K activity. Bands were quantified by LICOR odyssey software and the ratio of pAKT to total AKT at indicated concentrations of PI3K inhibitor, LY294002 is shown below the blots with values relative to untreated WT cells. C. qRT-PCR analysis of SDF-1 mRNA levels following treatment with an AKT inhibitor at the indicated concentrations D. Western blotting analysis of GSK phosphorylation, indicative of AKT activity. A representative of 3 independent experiments is shown and P value of <0.05 vs. WT; *compared to untreated WT cells, # compared to untreated YF cells.
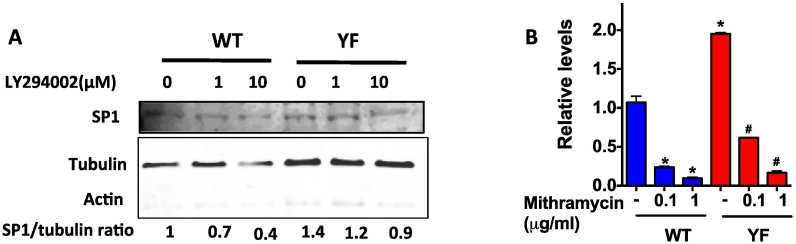
SDF-1 promoter harbors several Specificity protein-1 (Sp1) binding sites, and the requirement of Sp1 activation for SDF1 transcription was previously reported (Garcia-Moruja et al., 2005; Mendez-Ferrer et al., 2008; Schajnovitz et al., 2011). PI3K activity is an important modulator of Sp1 mediated transcription (Chu, 2012; Zhang et al., 2006). We examined the protein expression of the transcription factor Sp1. Sorted (CD45− Ter119− CD31− Sca1−) bone marrow cells were subjected to nuclear fractionation. Western blotting showed higher Sp1 protein in YF samples compared to WT nuclear lysates. Inhibition of PI3K lowered Sp1 levels in nuclei however, significant decrease in Sp1 nuclear levels was only observed at 10 μM LY294002 in YF cells (Fig. 3A). The enhanced nuclear translocation of Sp1 in YF cells was also confirmed by immunohistochemistry. Qualitatively, higher Sp1 protein levels were found in the nucleus of YF compared to WT cells. PI3K inhibition led to a decreased Sp1 nuclear localization in the nucleus of WT cells, but a decrease in staining intensity was not evident in YF cells (Suppl. Fig. 2).
Fig. 3.
Sp1 mediated transcription of SDF-1. A. Nuclear protein from cells treated with LY294002 for 3 h at different concentrations was subjected to Western blotting. Blot was probed with antibody for SP1. The blot was stripped and reprobed for actin (in cytoplasm) and tubulin (in nucleus) to show the relative absence of cytoplasmic proteins in the nuclear lysate. Sp1/tubulin ratio is shown below the Western blot. B. Cells were treated with Mithramycin (inhibits Sp1 binding to GC-rich DNA) at indicated concentrations for 3 h and SDF-1 mRNA levels were determined by qRT-PCR analysis. Values are shown as relative to untreated WT cells. * A representative of 3 independent experiments is shown and P value of <0.05 from student's t-test compared to untreated WT cells, # compared to untreated YF cells.
To confirm that Sp1 mediates increased SDF-1 transcription, cells were also treated with Mithramycin A, which blocks Sp1 binding to GC-rich regions on the SDF-1 promoter (Mendez-Ferrer et al., 2008; Choi et al., 2014). Mithramycin A treatment significantly blocked SDF-1 mRNA levels in WT and YF cells though the extent of inhibition was less pronounced in YF samples (Fig. 3B).
3.3. Migration of osteoclast precursors in response to SDF-1 stimulation was increased in YF mice
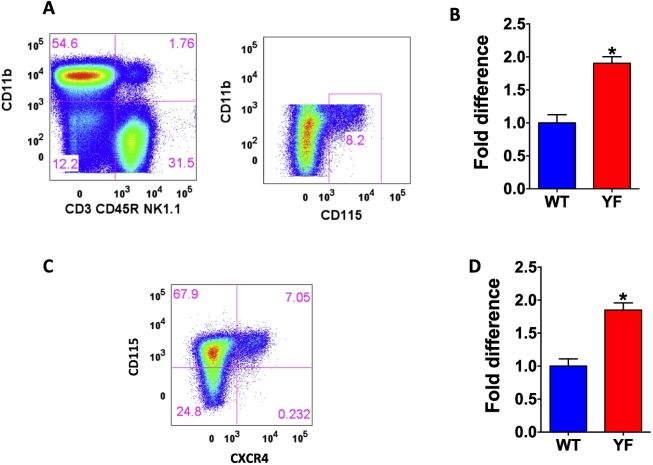
YF mice contain an increased number of osteoclasts, which are defective in bone resorptive function (Adapala et al., 2010a; Scanlon et al., 2015). We performed flow cytometric analysis to determine the number of osteoclast precursors in the bone marrow. Osteoclast precursors were identified as CD3− B220− NK1.1− CD11blo/− CD115hi cells (Jacquin et al., 2006), and percentage of osteoclast precursors was increased in YF bone marrow by 2-fold (Fig. 4A and B). The expression of SDF-1 receptor, CXCR4 was also increased in YF osteoclast precursors by 2-fold (Fig. 4 C and D).
Fig. 4.
Absence of Cbl-PI3K interaction results in increased number of osteoclast precursors expressing SDF-1 receptor, CXCR4 in the bone marrow of mice. Flow analysis of osteoclast precursors in the bone marrow. A. Total bone marrow from 8 week old male mice was gated for CD3- CD45R- NK1.1- cells, which express low or no CD11b and high levels of CD115 (c-Fms) were considered osteoclast precursors. B. Percentage of cells was calculated as the number in YF tissues relative to WT, and data pooled from independent experiments is shown (WT, n = 25; YF, n = 28). C. CD3- CD45R- NK1.1- CD11b lo/- CD115 hi cells were further gated for the expression of SDF-1 receptor CXCR4. D. CXCR4 positive osteoclast precursor population is shown (WT, n = 13; YF, n = 14). Percentage of cells was calculated as the number in YF tissues relative to WT, and data pooled from independent experiments is shown. * P value of <0.05 vs. WT.
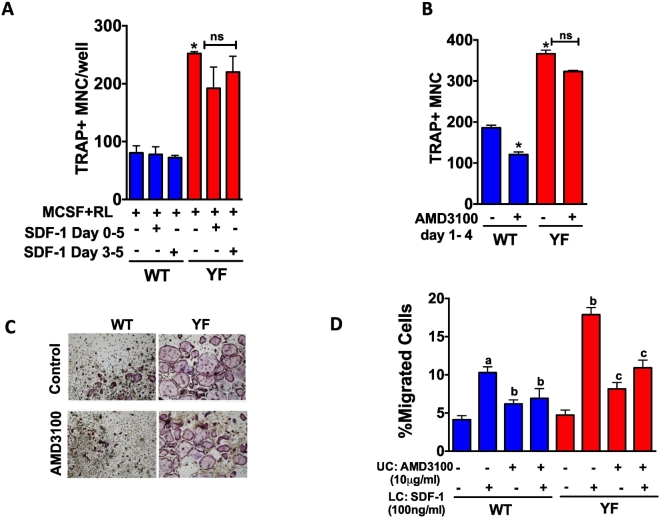
To understand if SDF-1 can influence osteoclast differentiation, the ability of WT and YF osteoclast precursors to differentiate into mature osteoclasts in response to SDF-1 was tested. Addition of SDF-1 to osteoclast differentiation medium either for all 5 days of culture or just 3–5 days did not alter the differentiation osteoclast precursors for either genotype (Fig. 5A). However, YF osteoclast precursors showed enhanced differentiation into mature osteoclasts as previously reported (Adapala et al., 2010a, Adapala et al., 2014a). While inhibition of SDF-1 binding to CXCR4 decreased osteoclast differentiation by 32% in WT cultures, treatment of YF cultures with the inhibitor did not influence the enhanced osteoclast formation (Fig. 5B and C) suggesting increased differentiation of YF osteoclasts is not mediated by SDF-1.
Fig. 5.
SDF-1 mediates increased migration but not differentiation of osteoclast precursors. Transmigration assay of osteoclast precursors in response to SDF-1. A. Osteoclast precursors were placed in the upper chamber of the transwell with or without AMD3100. SDF-1 was added to the lower chamber. Percentage of cells, which migrated to the lower chamber was quantified and data is expressed relative to WT cells without treatment in the absence of SDF-1 in the lower chamber. B. Osteoclast precursors were differentiated in the presence of MCSF (20 ng/ml) and RANKL (50 ng/ml) in the presence or absence of SDF-1 (100 ng/ml) added at indicated times. After 5 days of differentiation, cells were TRAP stained and TRAP positive cells with >3 nuclei were considered as osteoclasts. C. Osteoclast differentiation was performed in the presence or absence ofAMD3100 with SDF-1 treatment (100 ng/ml). Images of TRAP positive cells at 10× magnification are shown. D. Bar graph shows the number of multinucleated (>3 nuclei/cell) TRAP positive cells. In panel A, a: compared to untreated WT cells, b: compared to WT cells exposed to SDF-1 in lower chamber, c: compared to YF cells exposed to SDF-1 in lower chamber. In B, D * compared to WT cells without SDF-1 or AMD3100 treatment. A representative of 3 independent experiments is shown. * P value of <0.05 from ANOVA, Tukey's post hoc test was considered statistically significant.
We also analyzed the effect of SDF-1 on the migration of osteoclast precursors using transmigration assay. Both WT and YF osteoclast precursors showed comparable ability to migrate under basal conditions. In response to SDF-1, 11% of WT osteoclast precursors migrated compared to 17% of YF osteoclast precursors (Fig. 5D). Treatment with AMD3100 attenuated the migration response to SDF-1 for both genotypes.
4. Discussion
The PI3K-AKT signaling node regulates cell proliferation, differentiation, and migration of cells in the bone marrow environment. To our knowledge there are no previous studies that interrogate the direct impact of increased PI3K-AKT activity in regulating SDF1 production in the bone marrow milieu. We used the YF mice to understand the impact of PI3K activation on SDF-1 expression for several reasons. The majority of the studies examining how PI3K signaling governs bone homeostasis were done using knock out mice (Gyori et al., 2014; Shinohara et al., 2012). However, the deletion of PI3K genes often alters expression of non-targeted family members, precluding accurate phenotypic analysis (Bi et al., 1999; Fruman et al., 2000; Vanhaesebroeck et al., 2005). Furthermore, complete gene abrogation or overexpression of regulatory or catalytic subunits fails to take into account the spatio-temporal protein-protein interactions between the regulatory and catalytic subunits that regulate PI3K enzymatic activity. Additionally, the approach of generating point mutation(s) in the catalytic or the regulatory subunits results in malignant phenotypes (Burke and Williams, 2015; Jaiswal et al., 2009) rendering them undesirable for the study of homeostasis.
The primary function of the p85 regulatory subunit is to bind and stabilize the p110 subunit thereby modulating PI3K activity (Cantley, 2002). While establishing the function of Cbl in bone resorption (Adapala et al., 2014a; Chiusaroli et al., 2003; Nakajima et al., 2009), we identified a unique function of Cbl through the requirement of a tyrosine 737 for its interaction with the SH2 domain of the p85 regulatory subunit of PI3K (Songyang and Cantley, 1995). To study the impact of this interaction, YF mice, a global knock-in mouse model in which the tyrosine 737 was substituted to phenylalanine (Molero et al., 2006) was used. Our characterization of YF mice revealed that lack of Cbl-PI3K interaction results in increased PI3K-AKT signaling and increased level of SDF-1. This in turn perturbs bone homeostasis, thereby affecting both osteoclast and osteoblast differentiation and function (Adapala et al., 2010a, Adapala et al., 2010b, Adapala et al., 2014a, Adapala et al., 2014b; Brennan et al., 2011).
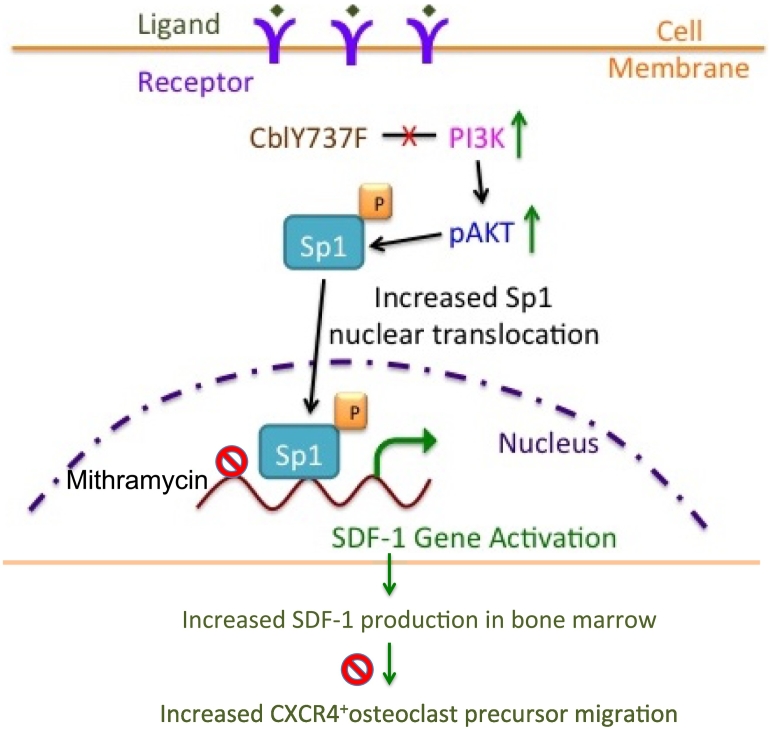
In this report we found that PI3K activity regulates SDF-1 production in CAR cells by modulating the Sp1 transcription factor. A schematic representation of the proposed mechanism is shown in Fig. 6. Several cell types including, osteoprogenitors, CAR cells (Greenbaum et al., 2013; Omatsu et al., 2010; Sugiyama et al., 2006), and CD31+ endothelial cells (Greenbaum et al., 2013; Sugiyama and Nagasawa, 2012; Mendez-Ferrer et al., 2010) produce SDF-1. We found that upregulation of PI3K-AKT activity led to increased SDF-1 expression by CAR cells. Others have also shown that CAR cells are critical source of SDF-1 and are responsible for maintaining hematopoietic niches (Sugiyama et al., 2018). While we found that in CD31+ cells SDF-1 expression was similar between the WT and YF cells, we cannot rule out that other cell sources, which produce SDF-1 and participate in hematopoiesis, may also contribute expression in YF mice. It is possible that increased SDF-1 can promote the proliferation of bone marrow stromal cells (Kortesidis et al., 2005), and enhanced SDF-1 levels might also be responsible for the increase in the numbers of CAR cells in the bone marrow of YF mice.
Fig. 6.
Proposed model depicting the role of PI3K/AKT/Sp1 axis on SDF-1 expression in CAR cells and the effect of increased SDF-1 levels on osteoclast precursor migration in response to increased PI3K activation. Loss of Cbl-PI3K interaction results in increased PI3K activity, which leads to increased phosphorylation of AKT. Sp1, an important substrate of PI3K is activated, translocated to the nucleus and binds to the SDF-1 promoter regions to activate SDF-1 transcription in CAR cells. Sp1 binding to SDF-1 promoter regions is inhibited by Mithramycin treatment resulting in decreased SDF-1 transcription to a lesser extent in YF cells compared to wild type cells due to increased Sp1 activation in YF cells. Increased SDF-1 gene expression leads to increased SDF-1 protein levels, which stimulate migration of osteoclast precursors expressing SDF-1 receptor, CXCR4. AMD3100 blocks CXCR4 activation by SDF-1 and prevents osteoclast precursor migration, to a lesser extent in YF cells compared to wild type cells. Increased osteoclast precursor migration might lead to increased recruitment to the bone marrow milieu finally contributing to increased number of osteoclasts in YF mice lacking Cbl-PI3K interaction.
PI3K-AKT activity regulates activation of several transcription factors. We have previously reported that in YF mice enhanced AKT activity is responsible for upregulation of Sp7 activation, resulting in increased bone formation during fracture healing (Scanlon et al., 2017). Sp1 transcriptional activity is also dependent on PI3K activation (Chu, 2012; Zhang et al., 2006). Here we found that expression of Sp1, a transcription factor known to regulate SDF-1 transcription (Mendez-Ferrer et al., 2008; Schajnovitz et al., 2011), is up regulated in YF mice derived CAR cells. The SDF-1 promoter has several Sp1 binding sites (Garcia-Moruja et al., 2005) and Sp1 is known to mediate SDF-1 transcription (Markovic et al., 2015). Recent studies have shown that the transcription factor Forkhead box c1 (Foxc1) is involved in the development of CAR cells and the production of SDF-1 (Omatsu et al., 2014). The activity of Foxc1 involves the PI3K/AKT pathway (Huang et al., 2017), therefore it is likely that in addition to Sp1 other transcription factors including Foxc1 may also regulate SDF1 expression in YF bone marrow cells.
The basal levels of osteoclast precursor migration were similar in WT and YF cells. However, the migration of YF cells was increased by SDF-1 in a CXCR4 dependent manner. Since, Cbl-PI3K interaction is lost in YF cells, a parallel pathway such as activation of Focal Adhesion Kinase (FAK) via Protein Kinase C (PKC), which is known to be a mediator of the SDF-1/CXCR4 pathway in cell migration (Glodek et al., 2007; Wang et al., 2000), might mediate the increased migration of YF cells. However, the precise molecular signaling mechanism responsible for increased SDF-1 mediated migration in YF cells remains to be elucidated.
Osteoclast precursors originate from the myeloid lineage of the hematopoietic stem cells (Jacquin et al., 2006; Arai et al., 1999). The proliferation of osteoclast precursors derived from YF bone marrow is similar to WT cells (Adapala et al., 2010a). Hence a different mechanism is responsible for the increased number of osteoclast precursors in YF bone marrow. Previous studies have shown that SDF-1 stimulates osteoclast precursor migration (Wright et al., 2005; Yu et al., 2003). Our results also indicate that in vitro SDF-1 stimulates increased migration of YF osteoclast precursors when compared to WT cells. SDF-1 did not alter osteoclast differentiation mediated by MCSF and RANKL a finding similar to other studies (Wright et al., 2005). Interestingly, differentiation of osteoclast precursors in the presence of MCSF and RANKL was inhibited by AMD3100, most likely due to suppression of motility needed for osteoclast precursor fusion. The lack of inhibitory effect of AMD3100 in YF cells is likely due to excessive responsiveness of YF osteoclast precursors to RANK signaling, as shown previously (Adapala et al., 2010a).
In conclusion, our study increases understanding of molecular mechanism involved in the regulation of the number of osteoclast precursors in mouse bone marrow. The role of PI3K/AKT/Sp1 axis in the regulation of SDF-1 production in CAR cells emphasizes the importance of the regulation of PI3K activity by Cbl. Our study also raises the possibility that CAR cells in the hematopoietic niche might not only regulate the hematopoietic composition, but also cellular composition by enhancing osteoclast precursor migration. Further studies are needed to provide direct evidence for the role of CAR cells in modulating osteoclast precursor numbers and establish the role of PI3K signaling in this process.
Conflict of interest
Authors have no conflict of interest.
Acknowledgments
Acknowledgments
This work was supported by National Institute of Health Grant (AR0550601) to A.S.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bonr.2019.100203.
Appendix A. Supplementary data
Supplementary figures
References
- Adapala N.S., Barbe M.F., Langdon W.Y., Nakamura M.C., Tsygankov A.Y., Sanjay A. The loss of Cbl-phosphatidylinositol 3-kinase interaction perturbs RANKL-mediated signaling, inhibiting bone resorption and promoting osteoclast survival. J. Biol. Chem. 2010;285:36745–36758. doi: 10.1074/jbc.M110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adapala N.S., Barbe M.F., Langdon W.Y., Tsygankov A.Y., Sanjay A. Cbl-phosphatidylinositol 3 kinase interaction differentially regulates macrophage colony-stimulating factor-mediated osteoclast survival and cytoskeletal reorganization. Ann. N. Y. Acad. Sci. 2010;1192:376–384. doi: 10.1111/j.1749-6632.2009.05346.x. [DOI] [PubMed] [Google Scholar]
- Adapala N.S., Barbe M.F., Tsygankov A.Y., Lorenzo J.A., Sanjay A. Loss of Cbl-PI3K interaction enhances osteoclast survival due to p21-Ras mediated PI3K activation independent of Cbl-b. J. Cell. Biochem. 2014;115:1277–1289. doi: 10.1002/jcb.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adapala N.S., Holland D., Scanlon V., Barbe M.F., Langdon W.Y., Tsygankov A.Y., Lorenzo J.A., Sanjay A. Loss of Cbl-PI3K interaction in mice prevents significant bone loss following ovariectomy. Bone. 2014;67:1–9. doi: 10.1016/j.bone.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai F., Miyamoto T., Ohneda O., Inada T., Sudo T., Brasel K., Miyata T., Anderson D.M., Suda T. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J. Exp. Med. 1999;190:1741–1754. doi: 10.1084/jem.190.12.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi L., Okabe I., Bernard D.J., Wynshaw-Boris A., Nussbaum R.L. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J. Biol. Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- Brennan T., Adapala N.S., Barbe M.F., Yingling V., Sanjay A. Abrogation of Cbl-PI3K interaction increases bone formation and osteoblast proliferation. Calcif. Tissue Int. 2011;89:396–410. doi: 10.1007/s00223-011-9531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J.E., Williams R.L. Synergy in activating class I PI3Ks. Trends Biochem. Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chiusaroli R., Sanjay A., Henriksen K., Engsig M.T., Horne W.C., Gu H., Baron R. Deletion of the gene encoding c-Cbl alters the ability of osteoclasts to migrate, delaying resorption and ossification of cartilage during the development of long bones. Dev. Biol. 2003;261:537–547. doi: 10.1016/s0012-1606(03)00299-9. [DOI] [PubMed] [Google Scholar]
- Choi E.S., Nam J.S., Jung J.Y., Cho N.P., Cho S.D. Modulation of specificity protein 1 by mithramycin A as a novel therapeutic strategy for cervical cancer. Sci. Rep. 2014;4:7162. doi: 10.1038/srep07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S. Transcriptional regulation by post-transcriptional modification–role of phosphorylation in Sp1 transcriptional activity. Gene. 2012;508:1–8. doi: 10.1016/j.gene.2012.07.022. [DOI] [PubMed] [Google Scholar]
- Engelman J.A., Luo J., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Feshchenko E.A., Langdon W.Y., Tsygankov A.Y. Fyn, Yes, and Syk phosphorylation sites in c-Cbl map to the same tyrosine residues that become phosphorylated in activated T cells. J. Biol. Chem. 1998;273:8323–8331. doi: 10.1074/jbc.273.14.8323. [DOI] [PubMed] [Google Scholar]
- Feshchenko E.A., Shore S.K., Tsygankov A.Y. Tyrosine phosphorylation of C-Cbl facilitates adhesion and spreading while suppressing anchorage-independent growth of V-Abl-transformed NIH3T3 fibroblasts. Oncogene. 1999;18:3703–3715. doi: 10.1038/sj.onc.1202672. [DOI] [PubMed] [Google Scholar]
- Foukas L.C., Okkenhaug K. Gene-targeting reveals physiological roles and complex regulation of the phosphoinositide 3-kinases. Arch. Biochem. Biophys. 2003;414:13–18. doi: 10.1016/s0003-9861(03)00177-2. [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Mauvais-Jarvis F., Pollard D.A., Yballe C.M., Brazil D., Bronson R.T., Kahn C.R., Cantley L.C. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat. Genet. 2000;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- Garcia-Moruja C., Alonso-Lobo J.M., Rueda P., Torres C., Gonzalez N., Bermejo M., Luque F., Arenzana-Seisdedos F., Alcami J., Caruz A. Functional characterization of SDF-1 proximal promoter. J. Mol. Biol. 2005;348:43–62. doi: 10.1016/j.jmb.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Glodek A.M., Le Y., Dykxhoorn D.M., Park S.Y., Mostoslavsky G., Mulligan R., Lieberman J., Beggs H.E., Honczarenko M., Silberstein L.E. Focal adhesion kinase is required for CXCL12-induced chemotactic and pro-adhesive responses in hematopoietic precursor cells. Leukemia. 2007;21:1723–1732. doi: 10.1038/sj.leu.2404769. [DOI] [PubMed] [Google Scholar]
- Grassi F., Cristino S., Toneguzzi S., Piacentini A., Facchini A., Lisignoli G. CXCL12 chemokine up-regulates bone resorption and MMP-9 release by human osteoclasts: CXCL12 levels are increased in synovial and bone tissue of rheumatoid arthritis patients. J. Cell. Physiol. 2004;199:244–251. doi: 10.1002/jcp.10445. [DOI] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronthos S., Zannettino A.C. The role of the chemokine CXCL12 in osteoclastogenesis. Trends Endocrinol Metab. 2007;18(3):108–113. doi: 10.1016/j.tem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Gyori D., Csete D., Benko S., Kulkarni S., Mandl P., Dobo-Nagy C., Vanhaesebroeck B., Stephens L., Hawkins P.T., Mocsai A. The phosphoinositide 3-kinase isoform PI3Kbeta regulates osteoclast-mediated bone resorption in humans and mice. Arthritis Rheumatol. 2014;66:2210–2221. doi: 10.1002/art.38660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Huang Z., Fan Y., He L., Ye M., Shi K., Ji B., Huang J., Wang Y., Li Q. FOXC1 promotes proliferation and epithelial-mesenchymal transition in cervical carcinoma through the PI3K-AKT signal pathway. Am. J. Transl. Res. 2017;9:1297–1306. [PMC free article] [PubMed] [Google Scholar]
- Jacquin C., Gran D.E., Lee S.K., Lorenzo J.A., Aguila H.L. Identification of multiple osteoclast precursor populations in murine bone marrow. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2006;21:67–77. doi: 10.1359/JBMR.051007. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring P, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–74. [DOI] [PMC free article] [PubMed]
- Kortesidis A., Zannettino A., Isenmann S., Shi S., Lapidot T., Gronthos S. Stromal-derived factor-1 promotes the growth, survival, and development of human bone marrow stromal stem cells. Blood. 2005;105:3793–3801. doi: 10.1182/blood-2004-11-4349. [DOI] [PubMed] [Google Scholar]
- Ma Q., Jones D., Springer T.A. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999;10:463–471. doi: 10.1016/s1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- Markovic J., Uskokovic A., Grdovic N., Dinic S., Mihailovic M., Jovanovic J.A., Poznanovic G., Vidakovic M. Identification of transcription factors involved in the transcriptional regulation of the CXCL12 gene in rat pancreatic insulinoma Rin-5F cell line. Biochem. Cell Biol. 2015;93:54–62. doi: 10.1139/bcb-2014-0104. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Lucas D., Battista M., Frenette P.S. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Sanjay A., Neff L., Tanaka S., Horne W.C., Baron R. Src kinase activity is essential for osteoclast function. J. Biol. Chem. 2004;279:17660–17666. doi: 10.1074/jbc.M311032200. [DOI] [PubMed] [Google Scholar]
- Molero J.C., Turner N., Thien C.B., Langdon W.Y., James D.E., Cooney G.J. Genetic ablation of the c-Cbl ubiquitin ligase domain results in increased energy expenditure and improved insulin action. Diabetes. 2006;55:3411–3417. doi: 10.2337/db06-0955. [DOI] [PubMed] [Google Scholar]
- Nagasawa T. CXC chemokine ligand 12 (CXCL12) and its receptor CXCR4. J. Mol. Med. 2014;92:433–439. doi: 10.1007/s00109-014-1123-8. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Nakajima T., Tachibana K., Iizasa H., Bleul C.C., Yoshie O., Matsushima K., Yoshida N., Springer T.A., Kishimoto T. Molecular cloning and characterization of a murine pre-B-cell growth-stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T., Hirota S., Tachibana K., Takakura N., Nishikawa S., Kitamura Y., Yoshida N., Kikutani H., Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Omatsu Y., Sugiyama T. Control of hematopoietic stem cells by the bone marrow stromal niche: the role of reticular cells. Trends Immunol. 2011;32:315–320. doi: 10.1016/j.it.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Nakajima A., Sanjay A., Chiusaroli R., Adapala N.S., Neff L., Itzsteink C., Horne W.C., Baron R. Loss of Cbl-b increases osteoclast bone-resorbing activity and induces osteopenia. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2009;24:1162–1172. doi: 10.1359/JBMR.090205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu Y., Sugiyama T., Kohara H., Kondoh G., Fujii N., Kohno K., Nagasawa T. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Omatsu Y., Seike M., Sugiyama T., Kume T., Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- Scanlon V., Soung do Y., Adapala N.S., Morgan E., Hansen M.F., Drissi H., Sanjay A. Role of Cbl-PI3K interaction during skeletal remodeling in a murine model of bone repair. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon V., Walia B., Yu J., Hansen M., Drissi H., Maye P., Sanjay A. Loss of Cbl-PI3K interaction modulates the periosteal response to fracture by enhancing osteogenic commitment and differentiation. Bone. 2017;95:124–135. doi: 10.1016/j.bone.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schajnovitz A, Itkin T, D'Uva G, Kalinkovich A, Golan K, Ludin A, Cohen D, Shulman Z, Avigdor A, Nagler A, et al. CXCL12 secretion by bone marrow stromal cells is dependent on cell contact and mediated by connexin-43 and connexin-45 gap junctions. Nat. Immunol. 2011;12:391–8. [DOI] [PubMed]
- Shinohara M, Nakamura M, Masuda H, Hirose J, Kadono Y, Iwasawa M, Nagase Y, Ueki K, Kadowaki T, Sasaki T, et al. Class IA phosphatidylinositol 3-kinase regulates osteoclastic bone resorption through protein kinase B-mediated vesicle transport. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2012;27:2464–75. [DOI] [PubMed]
- Songyang Z., Cantley L.C. Recognition and specificity in protein tyrosine kinase-mediated signaling. Trends Biochem. Sci. 1995;20:470–475. doi: 10.1016/s0968-0004(00)89103-3. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Nagasawa T. Bone marrow niches for hematopoietic stem cells and immune cells. Inflamm. Allergy Drug Targets. 2012;11:201–206. doi: 10.2174/187152812800392689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Omatsu Y., Nagasawa T. Niches for hematopoietic stem cells and immune cell progenitors. Int. Immunol. 2018;31:5–11. doi: 10.1093/intimm/dxy058. [DOI] [PubMed] [Google Scholar]
- Tokoyoda K., Egawa T., Sugiyama T., Choi B.I., Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L.C. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Wang J., Knaut H. Chemokine signaling in development and disease. Development. 2014;141:4199–4205. doi: 10.1242/dev.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.F., Park I.W., Groopman J.E. Stromal cell-derived factor-1alpha stimulates tyrosine phosphorylation of multiple focal adhesion proteins and induces migration of hematopoietic progenitor cells: roles of phosphoinositide-3 kinase and protein kinase C. Blood. 2000;95:2505–2513. [PubMed] [Google Scholar]
- Wright L.M., Maloney W., Yu X., Kindle L., Collin-Osdoby P., Osdoby P. Stromal cell-derived factor-1 binding to its chemokine receptor CXCR4 on precursor cells promotes the chemotactic recruitment, development and survival of human osteoclasts. Bone. 2005;36:840–853. doi: 10.1016/j.bone.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Yu X., Huang Y., Collin-Osdoby P., Osdoby P. Stromal cell-derived factor-1 (SDF-1) recruits osteoclast precursors by inducing chemotaxis, matrix metalloproteinase-9 (MMP-9) activity, and collagen transmigration. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2003;18:1404–1418. doi: 10.1359/jbmr.2003.18.8.1404. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liao M., Dufau M.L. Phosphatidylinositol 3-kinase/protein kinase Czeta-induced phosphorylation of Sp1 and p107 repressor release have a critical role in histone deacetylase inhibitor-mediated derepression [corrected] of transcription of the luteinizing hormone receptor gene. Mol. Cell. Biol. 2006;26:6748–6761. doi: 10.1128/MCB.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Guo R., Schwarz E.M., Boyce B.F., Xing L. TNF inhibits production of stromal cell-derived factor 1 by bone stromal cells and increases osteoclast precursor mobilization from bone marrow to peripheral blood. Arthritis Res. Ther. 2008;10:R37. doi: 10.1186/ar2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Liang G., Huang Z., Doty S.B., Boskey A.L. Conditional inactivation of the CXCR4 receptor in osteoprecursors reduces postnatal bone formation due to impaired osteoblast development. J. Biol. Chem. 2011;286:26794–26805. doi: 10.1074/jbc.M111.250985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures