Figure 1.
scRNA-Seq Analysis Reveals Cell Lineages in Control and Nicotine-Exposed Embryoid Bodies
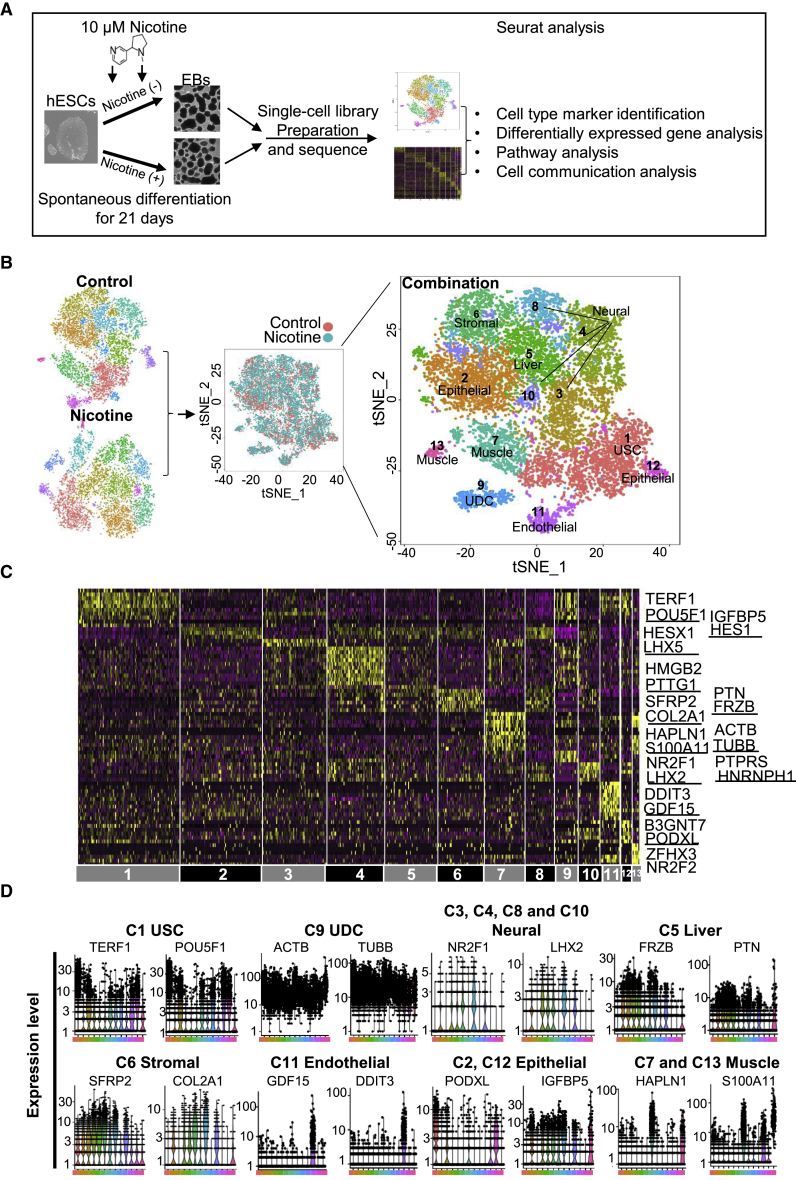
(A) Process flow diagram of scRNA-seq analysis on hESC differentiation. Single cells were collected from two independent EB differentiation experiments from day 21 EBs (nicotine-exposed versus control) and were prepared by single-cell barcoded droplets and chemicals from 10× Genomics. Bioinformatics data were processed using Seurat. Cell-type marker, differentially expressed gene, cell communication, and pathway analyses were performed to investigate the effects of nicotine exposure on hESC differentiation.
(B) Separated (left) and combined (middle and right) t-SNE plots of single cells from control and nicotine-exposed EBs. We defined six main types of progenitor cells in day 21 EBs, including muscle progenitor cells (clusters 3 and 13), liver progenitor cells (cluster 5), neural progenitor cells (clusters 3, 4, 8, and 11), stromal progenitor cells (cluster 6), epithelial progenitor cells (clusters 2 and 12), and endothelial progenitor cells (cluster 11). In addition, undifferentiated stem-like cells (USCs) (cluster 1) and undetermined cells (UDCs) (cluster 9) were also identified.
(C) Heatmap showing the expression pattern of top 10 differential genes in each cell type. Representative differential genes for each cell type are listed on the right side. The complete lists of differential genes for each cell type are listed in Table S3.
(D) Violin plots show the expression level distributions of marker genes across cell types.