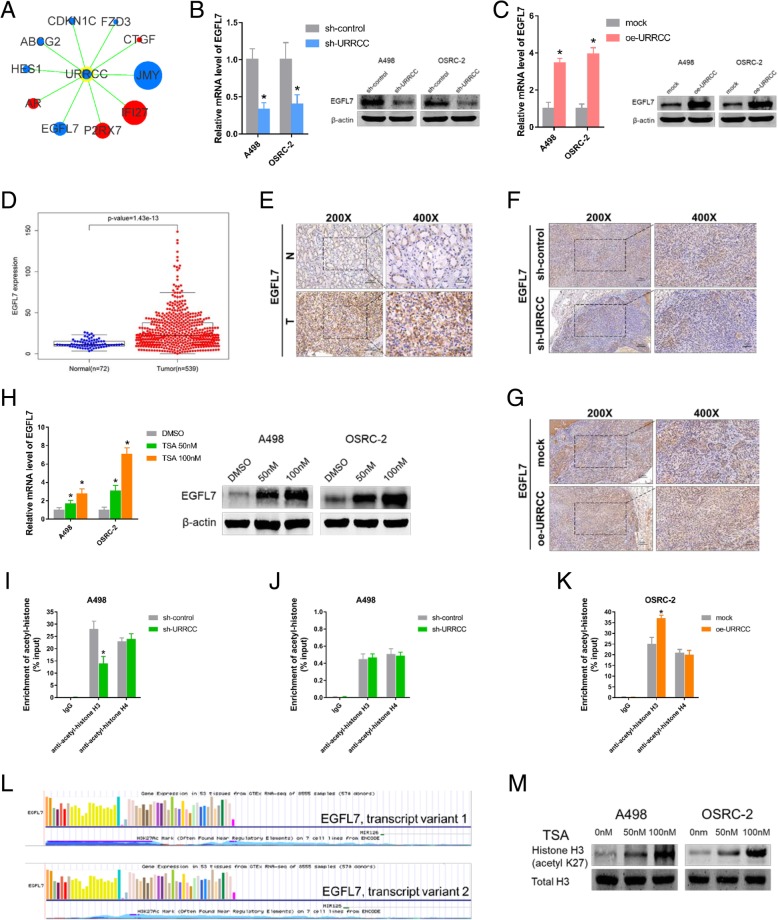
Fig. 4.
URRCC enhances EGFL7 level by mediating histone H3 acetylation of EGFL7 promoter. a: URRCC subnetwork according to the result of Target mRNA PCR Array. Genes colored in blue are down-regulated genes after that cells were transfected with sh-URRCC. Genes colored in red are up-regulated genes after that cells were transfected with sh-URRCC. The size of the gene round represents the fold change. b: The mRNA and protein levels of EGFL7 were detected by qRT-PCR and WB assays in A498 and OSRC-2 cells after transfection with sh-control or sh-URRCC. c: The mRNA and protein levels of EGFL7 were detected by qRT-PCR and WB assays in A498 and OSRC-2 cells after transfection with mock or oe-URRCC. d: EGFL7 expression in ccRCC and normal samples from TCGA KIRC dataset. e: Representative EGFL7 IHC staining of ccRCC tissues compared to paired normal renal tissues (200×, 400×). Black scale bar represents 100 μm (200×) or 50 μm (400×). f and g: Representative EGFL7 IHC staining of xenograft tumors from sh-control, sh-URRCC, mock, and oe-URRCC groups (200×, 400×). Black scale bar represents 100 μm (200×) or 50 μm (400×). h: qRT-PCR and WB analysis of EGFL7 in A498 and OSRC-2 cells treated with DMSO or trichostatin A (TSA) (50 nM or 100 nM) for 72 h (n = 3). i: ChIP analyses of A498 cells transfected with sh-control or sh-URRCC were conducted on the EGFL7 promoter regions using anti-acetyl-histone H3 and anti-acetyl-histone H4. Enrichment was determined relative to input controls. The data are the means ± standard deviations of three independent biological replicates. j: ChIP analyses of A498 cells transfected with sh-control or sh-URRCC were conducted on the GAPDH promoter regions using anti-acetyl-histone H3 and anti-acetyl-histone H4. Enrichment was determined relative to input controls. k: ChIP analyses of OSRC-2 cells transfected with mock or oe-URRCC were conducted on the EGFL7 promoter regions using anti-acetyl-histone H3 and anti-acetyl-histone H4. Enrichment was determined relative to input controls. The data are the means ± standard deviations of three independent biological replicates. l: EGFL7 promoter region was enriched with H3K27Ac histone mark presented with UCSC data. m: WB analysis of H3K27ac and total H3 in A498 and OSRC-2 cells treated with DMSO or TSA, β-actin was used as a loading control