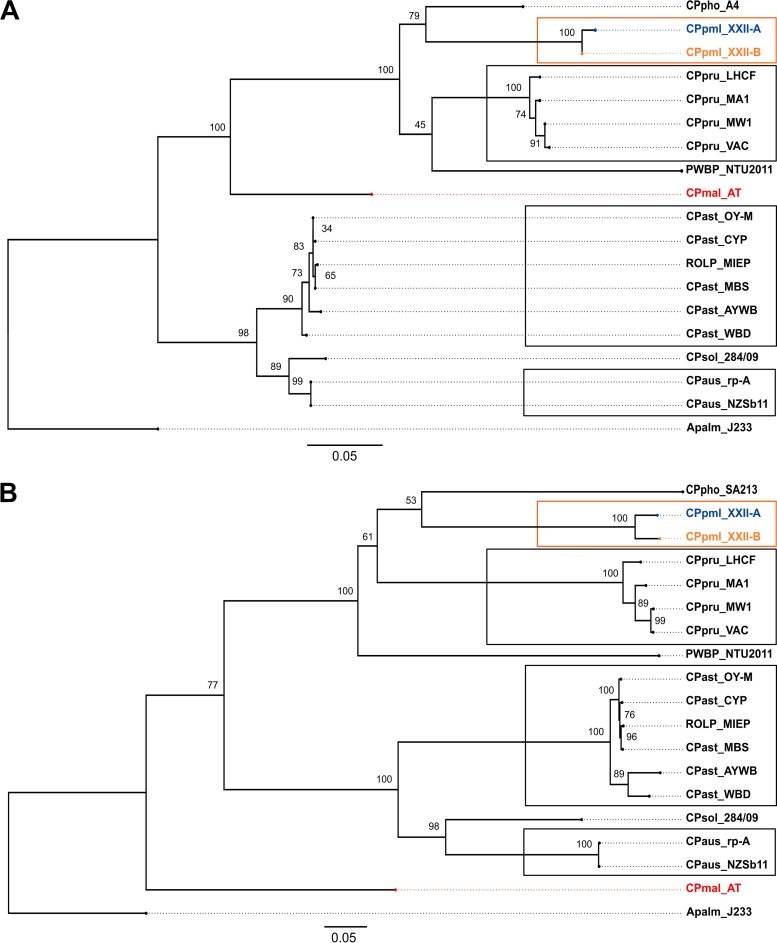
FIG 2.
Phylogenetic relationship of 18 Candidatus phytoplasmas. A rooted maximum likelihood tree was calculated for the 16S rRNA gene sequences (A) and for 3,756-bp concatenated DNA sequences of the dnaC, leuS, gyrB, rsmI, lpd, secA, and rplV housekeeping genes with the recombinant region removed (B). The codes used are as follows: CPmal_AT (“Ca. Phytoplasma mali” strain AT [CU469464.1]), CPaus_rp-A (“Ca. Phytoplasma australiense” [NC_010544.1]), CPast_OY-M (“Ca. Phytoplasma asteris” strain onion yellows phytoplasma OY-M [NC_005303.2]), CPpru_MW1 (“Ca. Phytoplasma pruni” strain milkweed yellows-MW1 [AKIL00000000.1]). CPpru_MA1 (“Ca. Phytoplasma pruni” strain Italian clover phyllody MA1 [AKIM00000000.1]), CPpru_VAC (“Ca. Phytoplasma pruni” strain vaccinium witches'-broom VAC [AKIN00000000.1]), PWBP_NTU2011 (peanut witches'-broom phytoplasma NTU2011 [AMWZ00000000.1]), CPaus_NZSb11 (“Ca. Phytoplasma australiense” strain strawberry lethal yellows phytoplasma NZSb11 [NC_021236.1]), CPsol_284/09 (“Ca. Phytoplasma solani” strain 284/09 [FO393427.1]), CPast_WBD (“Ca. Phytoplasma asteris” strain wheat blue dwarf [AVAO00000000.1]), CPpru_LHCF (“Ca. Phytoplasma pruni” strain CX [LHCF00000000.1]), and CPast_CYP (“Ca. Phytoplasma asteris” chrysanthemum yellows strain CYP [JSWH00000000.1]). CPast_AYWB (“Ca. Phytoplasma asteris” strain aster yellows witches broom [NC_007716.1]), CPpho_SA213 (“Ca. Phytoplasma phoenicum” [JPSQ00000000.1]) without recA, and CPho_A4 (AF515636), CPast_MBS (“Ca. Phytoplasma asteris” strain maize bushy stunt [NZ_CP015149.1]), and ROLP_MIEP (rice orange leaf phytoplasma LD1 [MIEP00000000.1]). Apalm_J233 (Acholeplasma palmae [NC_022538]) was used as an outgroup. “Candidatus Phytoplasma palmicola” is represented by one isolate (MZ11-005) from the 16SrXXII-A subgroup CPpml_XXII-A and by one isolate (GH04-009) from the 16SrXXII-B subgroup CPpml_XXII-B. Node values represent bootstrap test results with 500 replicates.