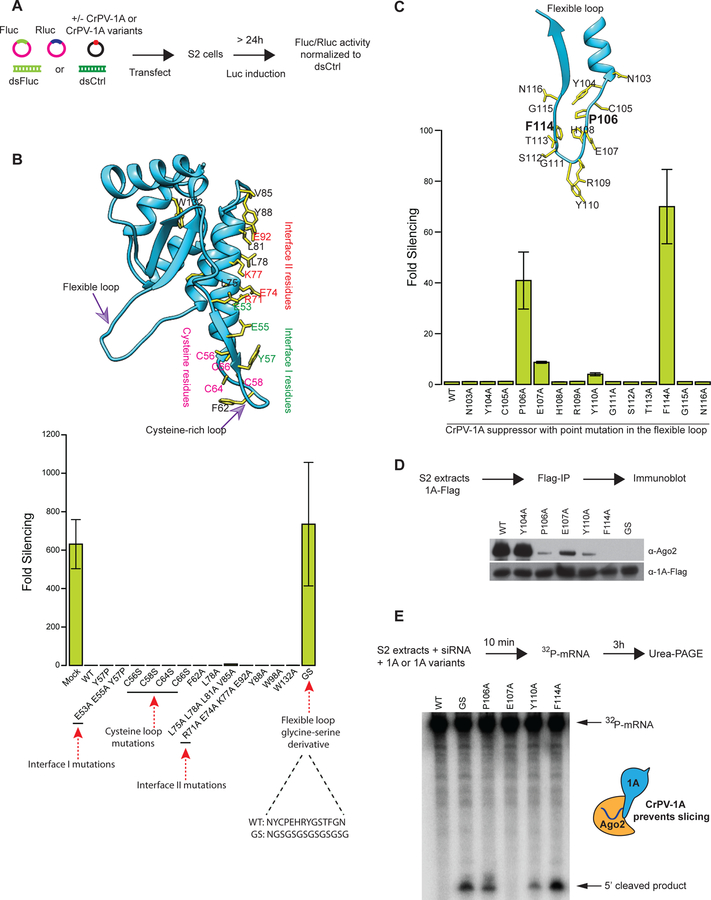
Fig 2: A flexible loop in CrPV-1A inhibits RNAi response in Drosophila cells.
(A) Schematic of a dual-luciferase reporter assay to screen for RNAi suppression (B) The CrPV-1A crystal structure showing amino acid residues selected for mutagenesis (yellow sticks) (top panel). The ability of these mutants to inhibit the RNAi response in S2 cells was tested in the RNAi reporter assay (bottom panel). (C) Residues (F114 and P106) in the flexible loop showing strongest inhibition of RNA silencing are highlighted in bold. The RNAi suppression data in (B) and (C) represent mean (±SD) of at least three independent experiments (n = 3) for each condition. (D) The affinity of CrPV-1A proteins for interactions with Ago-2 in S2 cells was probed by Flag-IP and Western blot analysis using antibodies directed against Drosophila Ago-2 protein. Expression levels of CrPV-1A proteins were probed with an anti-Flag antibody. One of three representative experiments is shown (n = 3). (E) S2 cell extracts were incubated with Fluc siRNA, radio-labeled capped Fluc RNA substrate, and purified CrPV-1A or variants. Slicing of the mRNA substrate was detected by denaturing PAGE and autoradiography. One of three representative experiments is shown (n = 3).