Fig 6: CrPV-1A hijacks cellular E3 ligase components.
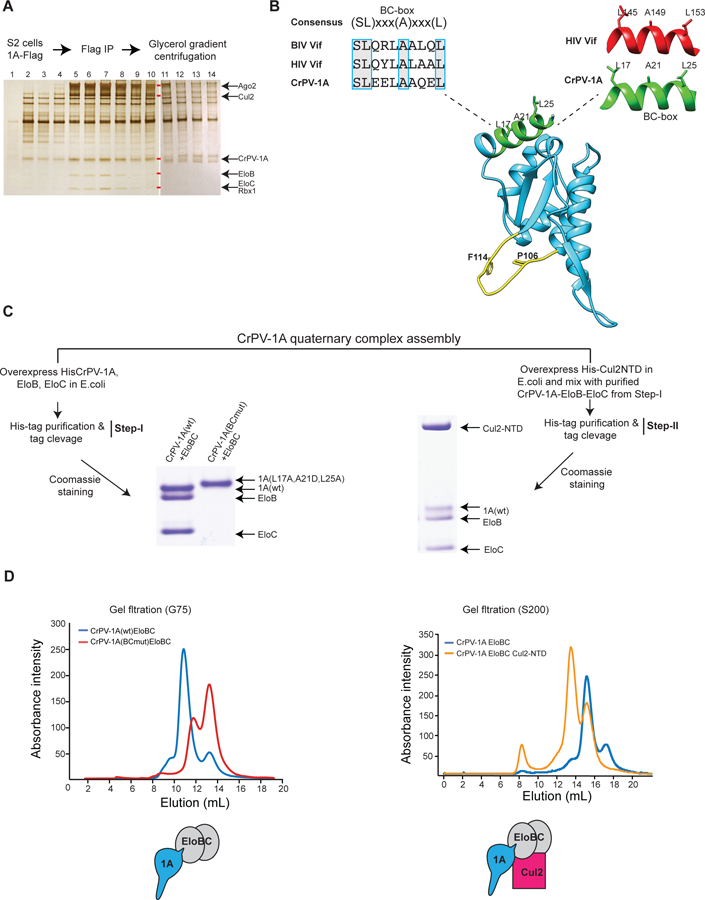
(A) Glycerol gradient fractions containing CrPV-1A associated proteins were analyzed using SDS-PAGE and silver staining. Gel bands corresponding to Ago-2, Cul2, EloB, EloC, and Rbx1 are indicated with red rectangles, and their identities were verified by mass spectrometry. After fraction 11, HSP27 was observed to run with similar electrophoretic mobility to CrPV-1A as confirmed by mass spectrometry. One of two representative silver stain gels is shown (n = 2). (B) Blue boxes highlight the consensus BC-box amino acid sequences of CrPV-1A, HIV Vif, and BIV Vif. Structural conservation of the BC-box motif for CrPV1A and HIV Vif is shown. The TALOS element residues (P106 and F114) and the BC-box motif are highlighted in yellow and green, respectively. (C) Assembly of CrPV-1A+EloB+EloC ternary and CrPV-1A+EloB+EloC+Cul2 quaternary complexes were examined by overexpression the proteins in E.coli, His-tag pull-down, and Coomassie blue staining. Elution patterns of the complexes was examined by Superdex-G75 and Superdex-200 column chromatography, respectively.
See also Fig S5
