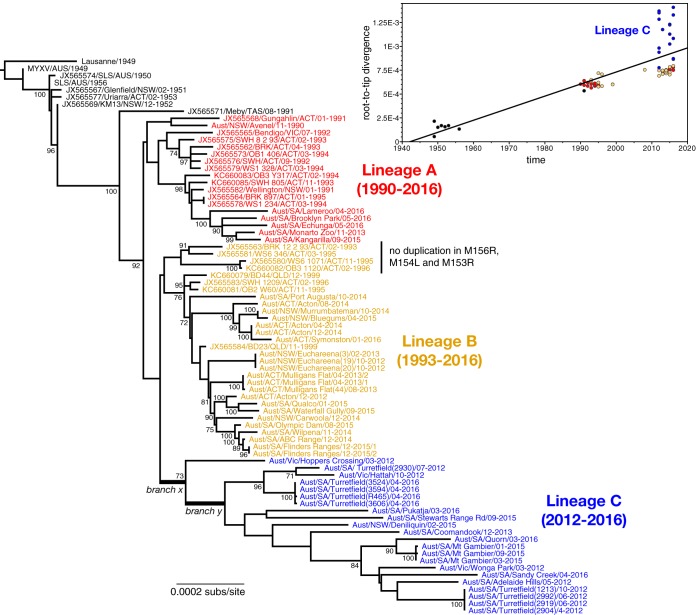
FIG 2.
Evolutionary history of MYXV in Australia. The tree depicts the phylogenetic relationships among all 78 complete genome sequences of MYXV sequenced to date. All viruses sampled from 1990 have been color coded and, with exception of the Meby strain sample in Tasmania in 1991, fall into three main lineages (denoted A, B, and C), with their range of sampling times shown. Two branches—denoted x and y—associated with the emergence of the rapidly evolving lineage C are indicated. All viruses within lineages B and C, aside from the four marked, possess a duplication in M156R, M154L, and part of M153R. The tree is rooted by the European Lausanne strain, and all branches are scaled according to the number of nucleotide substitutions per site (subs/site). All bootstrap values of >70% are shown. (Inset) Regression of root-to-tip genetic distances against the year of sampling for all available Australian isolates of MYXV. Each point is color coded according to the phylogenetic lineage. The recently sampled lineage C, which has experienced an elevation in evolutionary rate, is marked.