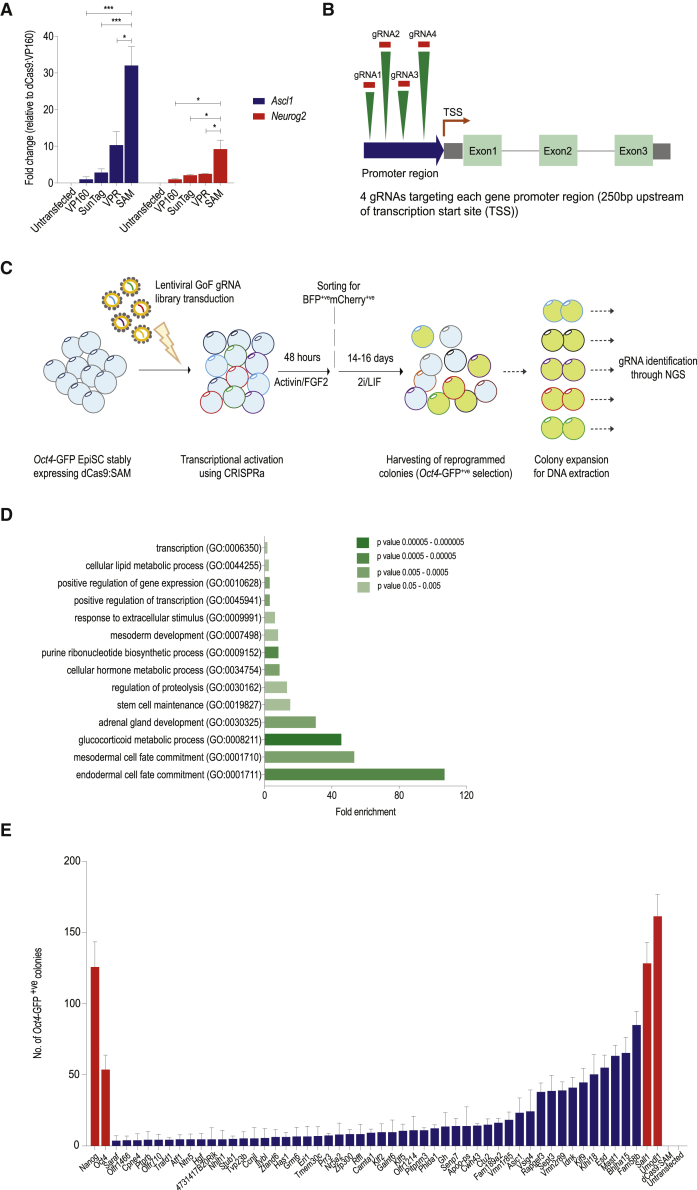
Figure 1.
GoF EpiSC Reprogramming Screening with CRISPRa and sgRNA Library
(A) Activation of Ascl1 and Neurog2 in HEK293 cells. Cells were transfected with one sgRNA per target and four different dCas9 versions. qRT-PCR normalized to Gapdh, fold change relative to dCas9:VP160 (mean of experimental triplicates ± SD, ∗p < 0.05; ∗∗∗p < 0.001).
(B) sgRNA design targeting gene promoters in the murine genome.
(C) Screening strategy in Oct4-GFP EpiSCs stably expressing dCas9:SAM, lentiviral transduction (MOI = 0.3) of the sgRNA library. Reprogramming in 2i/LIF for 14–16 days, after sorting for transduced cells. NGS identified candidate sgRNAs in Oct4-GFP+ve iPSC colonies.
(D) GOTOOLBox analysis of 142 genes identified in GoF screening. Pathways with fold change compared with reference; colors indicate p values.
(E) Validation of 54 genes including Nanog and Oct4 in dCas9:SAM-Oct4-GFP EpiSCs with single sgRNAs (Oct4-GFP+ve iPSC colonies, mean of 3 independent experiments ± SD).
See also Figures S1 and S2 and Tables S5, S6, and S7.