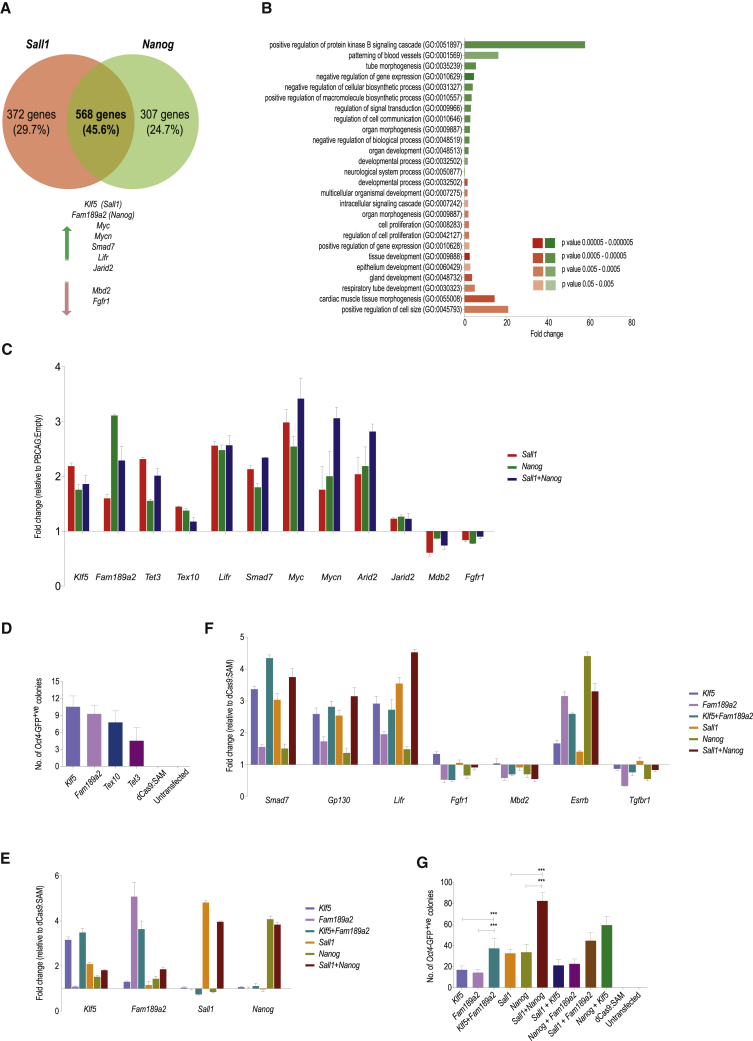
Figure 4.
RNA-Seq Identifies Potential Mechanisms of Reprogramming Mediated by Sall1 and Nanog
(A) Venn diagram of genes being differentially expressed in Sall1 and Nanog overexpressing cells (cutoff padj < 0.001). Upregulated (green arrow) and downregulated (red arrow) genes for further experiments were chosen from the overlap between Sall1 and Nanog, except Klf5 and Fam189a2.
(B) GOTOOLBox analysis of common regulated genes (fold change compared with reference, colors indicate p values).
(C) qRT-PCR validations of RNA-seq (24 h after transfection, normalized to Gapdh and relative to PBCAG:Empty; mean of 3 independent experiments ± SD).
(D) Reprogramming of Oct4-GFP EpiSCs via CRISPRa-mediated gene induction of Klf5, Fam189a2, Tex10, and Tet3 (Oct4-GFP+ colonies after 2i/LIF for 20 days; mean of 3 independent experiments ± SD).
(E) qRT-PCR for Klf5, Fam189a2, Sall1, and Nanog after CRISPRa-mediated induction of Klf5 and Fam189a2 (flow-sorted for sgRNA expression after 24 h, normalized to Gapdh and relative to dCas9:SAM; mean of 3 independent experiments ± SD).
(F) qRT-PCR expression levels of key regulators in JAK/STAT3 and TGF-β signaling (flow-sorted for sgRNA expression 24 h after changing to 2i/LIF media, 48 h after transfection, normalized to Gapdh and relative to dCas9:SAM; mean of 3 independent experiments ± SD).
(G) Reprogramming of Oct4-GFP EpiSCs via CRISPRa-mediated gene induction of Klf5, Fam189a2, Sall1, and Nanog (Oct4-GFP+ve colonies after 20 days in 2i/LIF; error bars represent mean of 3 independent experiments ± SD).