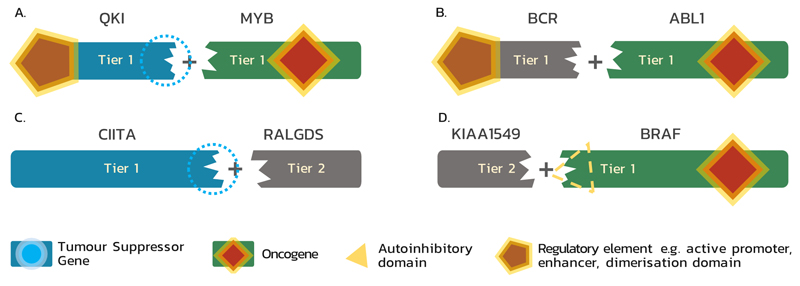
Figure 1. Functional classes of genes involved in fusions and their classification within the Cancer Gene Census.
(A) As a result of gene fusion, a tumour suppressor gene (TSG) may lose its suppressive abilities and a proto-oncogene may be transformed into an oncogene; e.g. QKI-MYB: in angiocentric glioma MYB is activated by truncation and the influence of the QKI enhancer; QKI loses its TSG function105. (B) Genes that, after fusion, upregulate an oncogene through donation of regulatory element (e.g. active promoter, enhancer, or activating domain) are included in Tier 1; e.g. BCR-ABL1: in this fusion (also known as the Philadelphia chromosome), BCR, which is neither TSG nor oncogene, simply provides an oligomerisation domain, which enables constitutive activation of ABL119. (C) A fusion partner which only deactivates a TSG by disrupting its sequence is classified as a Tier 2 Cancer Gene Census (CGC) gene if it is recurrently involved in a fusion; e.g. CIITA-RALGDS: in Hodgkin lymphoma this fusion results in the N-terminal part of CIITA, which loses its TSG function, fused in an out-of-frame fashion to RALGDS, which is neither TSG nor oncogene in this case 106. (D) A fusion may also result in hyperactivation of an oncogene due to loss of an autoinhibitory domain, which is replaced by a fusion partner; e.g. KIAA1549-BRAF: in pilocytic astrocytoma the N-terminal BRAF autoregulatory domain25 is lost in the fusion protein, resulting in constitutively active BRAF expressed under the control of the KIAA1549 promoter107. A fusion gene, such as KIAA1549, acting solely and recurrently through replacing a functional fragment of the other partner is included into Tier 2.