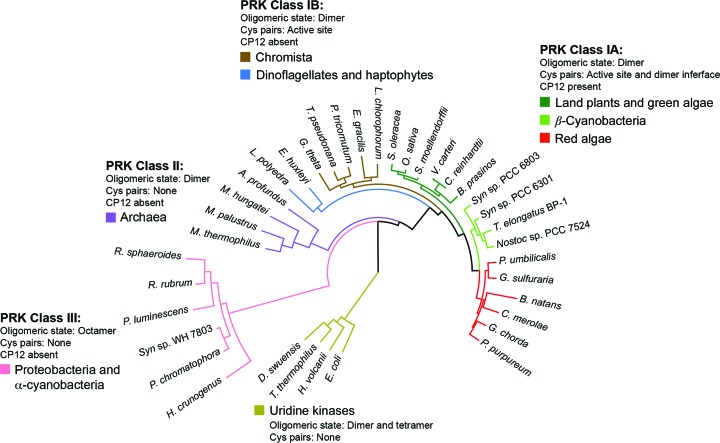
Figure 1.
Maximum-likelihood phylogenetic tree of PRK sequences. PRKs can be classified into three clear major groupings. Class I covers most photosynthetic organisms and can be further subdivided into two groups. Class IA contains vascular land plants, β-cyanobacteria and red algae, which have conserved cysteine pairs both in the active site and close to the C-terminus. Genomes with a class IA PRK also contain the CP12 adaptor protein. Class IB includes diatoms, dinoflagellates, haptophytes and other members of a vaguely defined ‘chromista’ clade. In contrast to class IA, most class IB PRKs lack the conserved cysteine residues close to the C-terminus and most of the source-organism genomes lack CP12. Class II enzymes are found in archaea and are also dimeric (Kono et al., 2017 ▸). Class III PRKs are distal to the other classes, do not feature redox regulation and have an octameric rather than a dimeric holoenzyme state. Uridine kinases, which belong to the same structural superfamily as PRK, form an outgroup in the phylogenetic tree.