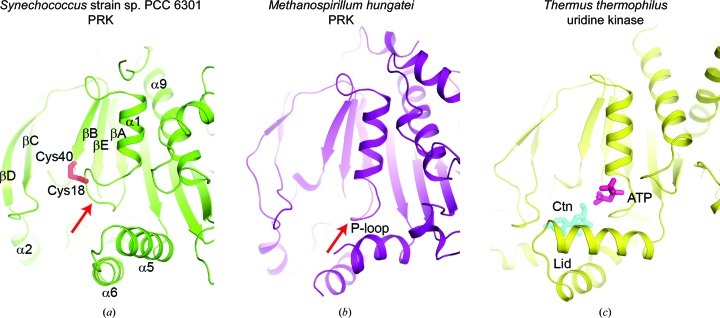
Figure 6.
Comparison of the active sites in Syn6301-PRK, MhPRK and uridine kinase. (a) Ribbon representation of Syn6301-PRK in the redox-blocked state. The Cys18–Cys40 disulfide bond is represented by red sticks. Secondary-structure elements are indicated. The red arrow indicates the Walker A motif (P-loop). (b) Ribbon representation of MhPRK (PDB entry 5b3f; Kono et al., 2017 ▸) in the apo state. The same view as in (a) is shown. The red arrow indicates the Walker A motif (P-loop). (c) Ribbon representation of uridine kinase from T. thermophilus (PDB entry 3w8r; Tomoike et al., 2013 ▸) in the substrate-bound state. The substrates cytidine (Ctn) and AMP-PCP (ATP) are presented as cyan and purple sticks, respectively. The helical hairpin acting as a lid above the substrate-binding sites is indicated.