Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Native | Osmate derivative | |
|---|---|---|
| PDB code | 6hzk | 6hzl |
| Resolution range (Å) | 30.0–2.40 (2.46–2.40) | 30.0–2.77 (2.84–2.77) |
| Completeness (%) | 99.8 | 99.9 |
| σ Cutoff | F > 0.000σ(F) | F > 0.000σ(F) |
| No. of reflections, working set | 27844 (1395) | 18412 (1343) |
| No. of reflections, test set | 1503 (108) | 1017 (66) |
| Final R work † | 0.222 (0.337) | 0.235 (0.338) |
| Final R free † | 0.257 (0.382) | 0.259 (0.414) |
| Cruickshank DPI | 0.3942 | 0.3879 |
| No. of non-H atoms | ||
| Protein | 4954 | 4954 |
| Ligand | 0 | 2 |
| Solvent | 21 | 1 |
| Total | 4975 | 4957 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.008 | 0.004 |
| Angles (°) | 1.218 | 0.897 |
| Average B factors (Å2) | ||
| Protein | 101‡ | 116§ |
| Ligand | — | 169 |
| Solvent | 64 | 58 |
| Ramachandran plot | ||
| Most favoured (%) | 97.7 | 98.5 |
| Allowed (%) | 1.8 | 1.5 |
| Outliers (%) | 0.5 | 0 |
R
work = 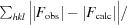
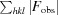 ; R
free is the R factor for a pre-selected subset (5%) of reflections that were not included in refinement.
; R
free is the R factor for a pre-selected subset (5%) of reflections that were not included in refinement.
The protein chains have very different average B factors: 69.4 Å for chain A and 136 Å for chain B.
The protein chains have very different average B factors: 79.7 Å for chain A and 156 Å for chain B.
