Figure 3.
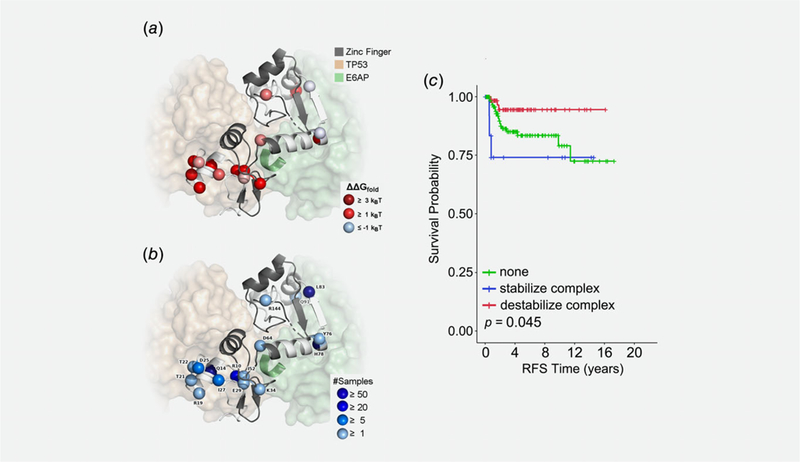
Structural and functional annotation of E6 variants and their association with outcome. We used existing experimental protein structures of E6 to calculate changes in stability for E6 alone and E6 in complex with TP53 (p53) and E6AP. (a) Cartoon of E6 with the residues in the two zinc finger domains colored gray and the other residues colored white. Spheres mark the variant sites, which are colored by their effects on folding stability of the protein complex. Proteins bound to E6 are shown in transparent surfaces, and the LXXLL motif of E6AP is shown in the cartoon. (b) The same sites are shown in spheres, now labeled by their WT residues and colored according to the number of samples they were observed. (c) Kaplan–Meier recurrence-free survival in ICC patients that is associated with HPV 16 nonsynonymous variants that were predicted to stabilize the E6–E6AP–p53 complex (blue line) or destabilize the E6–E6AP–p53 complex (red line) compared to ICC patients with no E6 variants (green line).
