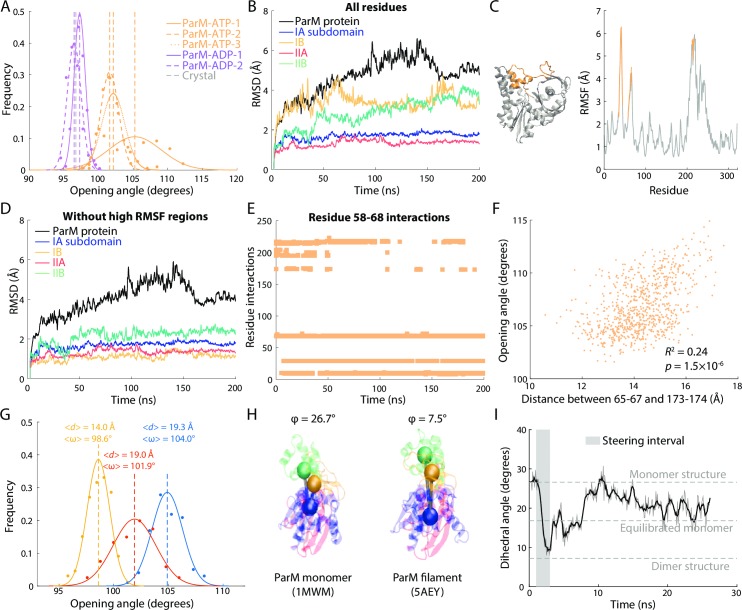
Fig 4. Loop in the IB domain drives ParM monomer opening.
(A) The opening angle of ADP-bound ParM monomers remained near the crystal structure value (gray dashed line), while that of ATP-bound ParM monomers consistently increased. Two simulations (ParM-ATP-2,3) consistently exhibited opening angles of ~102° after 50 ns and maintained that value, whereas in the other simulation (ParM-ATP-1), the opening angle increased beyond 105° after 100 ns and then eventually decreased to ~103° in the last 20 ns of the 200-ns simulation (S4A Fig). Dashed lines are mean values. Gray dashed line is the value in the crystal structure. (B) For the ATP-bound ParM simulation in which the opening angle increased beyond 105° (ParM-ATP-1), there were large increases in RMSD across the entire protein and in subdomains IIA and IIB. (C) RMSF analysis of single residue fluctuations during the simulation in (B) revealed two regions (residues 58–67 and 173–174, gold) with high RMSF values that were spatially proximal on the crystal structure. (D) For the simulation in (B), when excluding residues 58–67 and 173–174, the RMSDs of all four subdomains dropped to ~2 Å. Thus, these regions were responsible for the conformational variability in (B). (E) Interactions of the loop formed by residues 58–67 with residues 173–174 and 200–202 disappeared early in simulation ParM-ATP-1. Interactions were defined as a minimum distance between residues of <5 Å. (F) The distance (d) between residues 65–67 and 173–174 was highly correlated with the opening angle (ω) across the last 80 ns of the ParM-ATP-1 simulation. The p-value was computed with sampling corresponding to a time interval of 1 ns; for an interval of 3 ns, the p-value is 0.0037. (G) Steering of the distance between residues 65–67 and 173–174 tuned the opening angle in a distance-dependent manner. Dashed lines are mean values. (H) The dihedral angle of a ParM monomer crystal structure (PDB ID: 1MWM) was much higher than that of each subunit in a ParM filament crystal structure (PDB ID: 5AEY). (I) When the dihedral angle of a ParM-ATP monomer was steered to 7.5° (gray box) and then released, the angle stabilized at a value similar to unconstrained simulations (S4B Fig), indicating that ParM flattens upon polymerization.