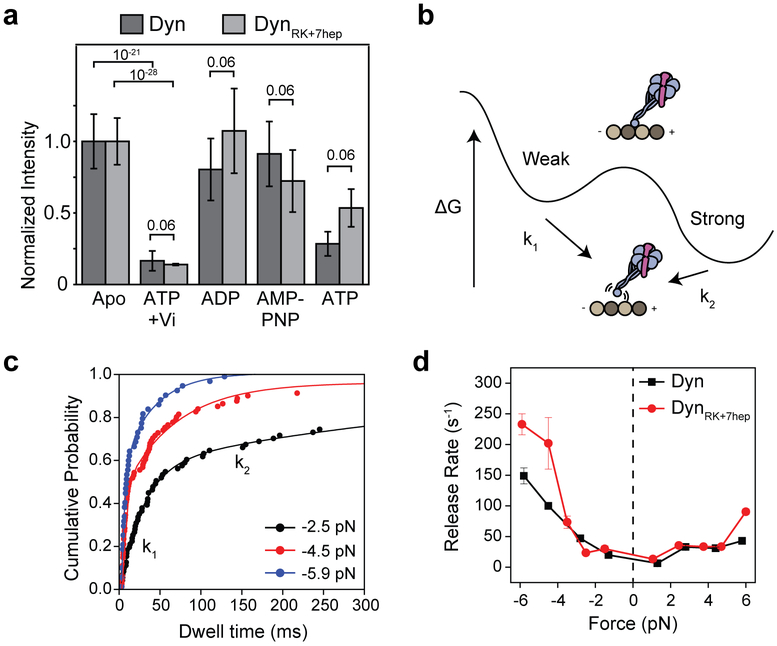
Extended Data Figure 8 ∣. Nucleotide and force-induced release of Dyn and DynRK+7hep monomers from MTs.
a, The normalized intensity of 100 nM GFP-tagged Dyn and DynRK+7hep monomers on sea urchin axonemes under given nucleotide conditions. Similar to Dyn, DynRK+7hep released from MTs in the ADP-Pi state, mimicked by ATP and vanadate (Vi, n = 40 axonemes from three independent measurements, mean ± s.d.). p-values are calculated from a two-sided t-test. b, A model of the dynein-MT interaction shows two distinct binding modes in the apo state, with k1 and k2 representing force-induced release rates from the weak and strong states, respectively. The slow rate (k2) represents strong binding of the motor to its tubulin binding site, whereas the fast rate (k1) represents transient or nonspecific interactions of the motor with the MT. c, Cumulative probability distributions (solid circles) of the MT-bound time of Dyn monomers at given force ranges. The release rates (k1 and k2) were calculated by a two-exponential-decay fit (solid curves). d, Calculated k1 values from the exponential fit (±95% confidence intervals) to the DynRK+7hep dwell time has similar force-dependence to Dyn. Each bin contains 120 dwells from two independent measurements.