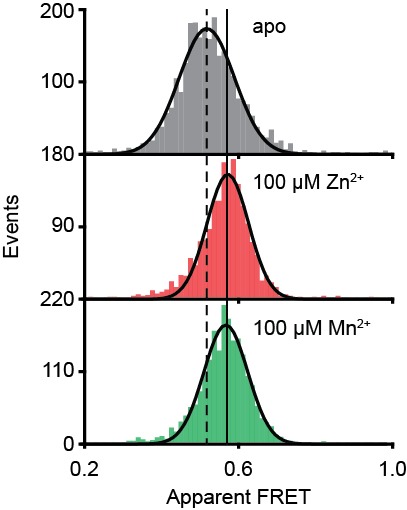
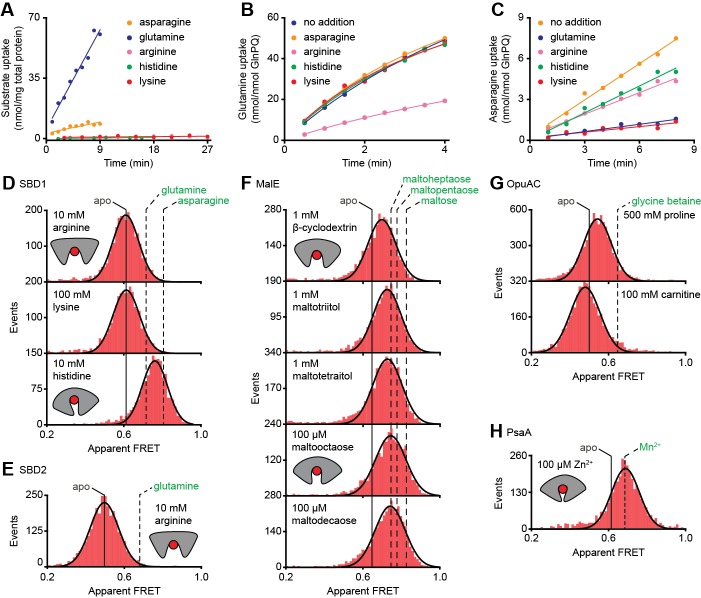
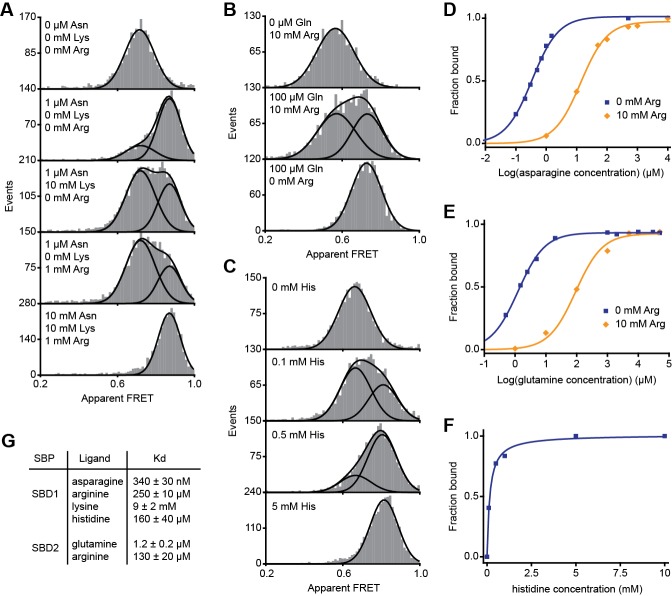
Figure 4. Substrate-specificity of GlnPQ and SBP conformations induced by non-cognate substrates.
(A) Time-dependent uptake [14C]-asparagine (5 μM), [14C]-glutamine (5 μM), [14C]-arginine (100 μM), [14C]-histidine (100 μM) and [3H]-lysine (100 μM) by GlnPQ in L. lactis GKW9000 complemented in trans with a plasmid for expressing GlnPQ; the final amino acid concentrations are indicated between brackets. Points are the data and the solid line a hyperbolic fit. Time-dependent uptake of glutamine (B) and asparagine (C) in proteoliposomes reconstituted with purified GlnPQ (see Materials and methods section). The final concentration of [14C]-glutamine and [14C]-asparagine was 5 μM, respectively; the amino acids indicated in the panel were added at a concentration of 5 mM. Solution-based apparent FRET efficiency histogram of SBD1(T159C/G87C) (D), SBD2(T369C/S451) (E), MalE(T36C/S352C) (F), OpuAC(V360C/N423C) (G) and PsaA(V76C/K237C) (H) in the presence of non-cognate (red bars) substrates as indicated. Bars are the data and solid line a Gaussian fit. The 95% confidence interval for the distribution mean is shown in Supplementary file 3. The interval center is indicated by vertical lines (solid and dashed).
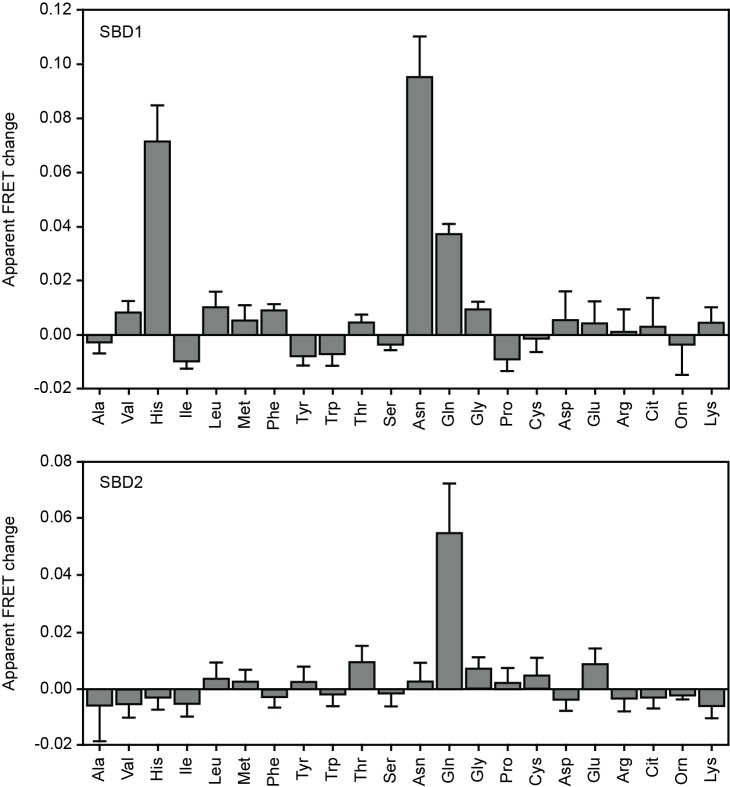
Figure 4—figure supplement 1. Substrate binding of SBD1 and SBD2 studied by ensemble FRET.
Figure 4—figure supplement 2. Non-cognate substrate binding by SBD1 and SBD2.
Figure 4—figure supplement 3. PsaA(E74C/K237C) conformational changes probed by smFRET.